Join Our Lab!
We are recruiting postdocs and graduate students for new and ongoing projects.
- Graduate Students
- Postdoctoral Associates
Graduate Students:
- Contact Tom Cooper to discuss rotation opportunities.
About the Lab

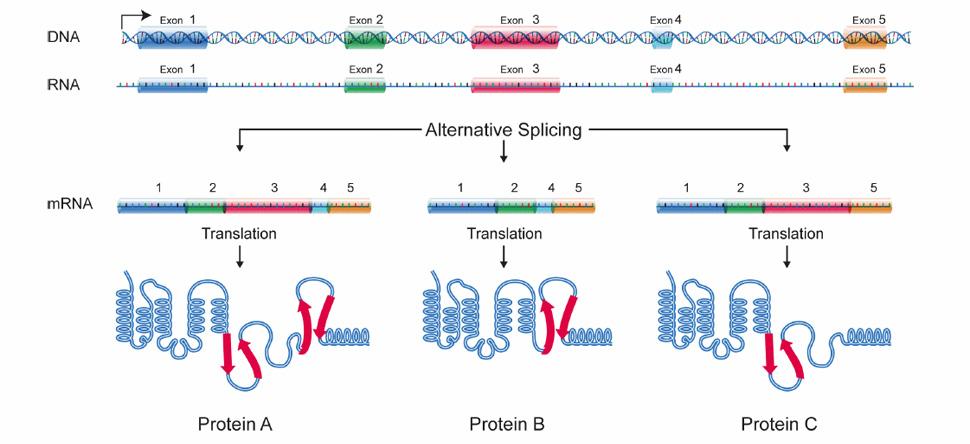
The realization that most human genes generate multiple mRNAs encoding divergent protein isoforms via alternative splicing and polyadenylation has revealed extensive regulation that remains to be explored.
Alternative pathways of RNA processing are regulated in response to signaling cues often coordinating expression of gene networks in response to physiological change including in development and disease.
We are interested in understanding the mechanisms and consequences of this regulation, from how RNA binding proteins and signaling pathways coordinate RNA processing networks to the functional consequences of the different protein isoforms that are expressed in different cell states.
We also investigate the pathogenic mechanisms of myotonic dystrophy, type 1 (DM1), an autosomal dominant neuromuscular disorder affecting multiple tissues including muscle, heart and the central nervous system. The pathogenic mechanism is disruption of developmentally regulated RNA processing, primarily alternative splicing, in which failure to express adult splicing patterns causes primary features of the disease.
The understanding of the molecular mechanisms of DM1 pathogenesis has led to development of several therapeutic approaches some of which are being testing using mouse models established in the lab.
These investigations utilize a combination of cell culture and genetic models including transgenic and knock out mouse lines for RNA binding proteins, CRISPR-derived mouse lines in which specific alternative exons are removed, and DM1 mouse models.
The overlapping areas of investigation in the lab lead to synergistic and collaborative interactions in which knowledge gained in one area fosters progress in the others.
Cooper Lab News
Rong-Chi’s paper describing rescue of the mouse heart model for myotonic dystrophy by muscleblind expression was published in the J. Clinical Investigation, 2025
Lathan defended his thesis and graduated 2025
Jesus Frias, Ph.D. joined our lab as a new postdoctoral fellow, 2024
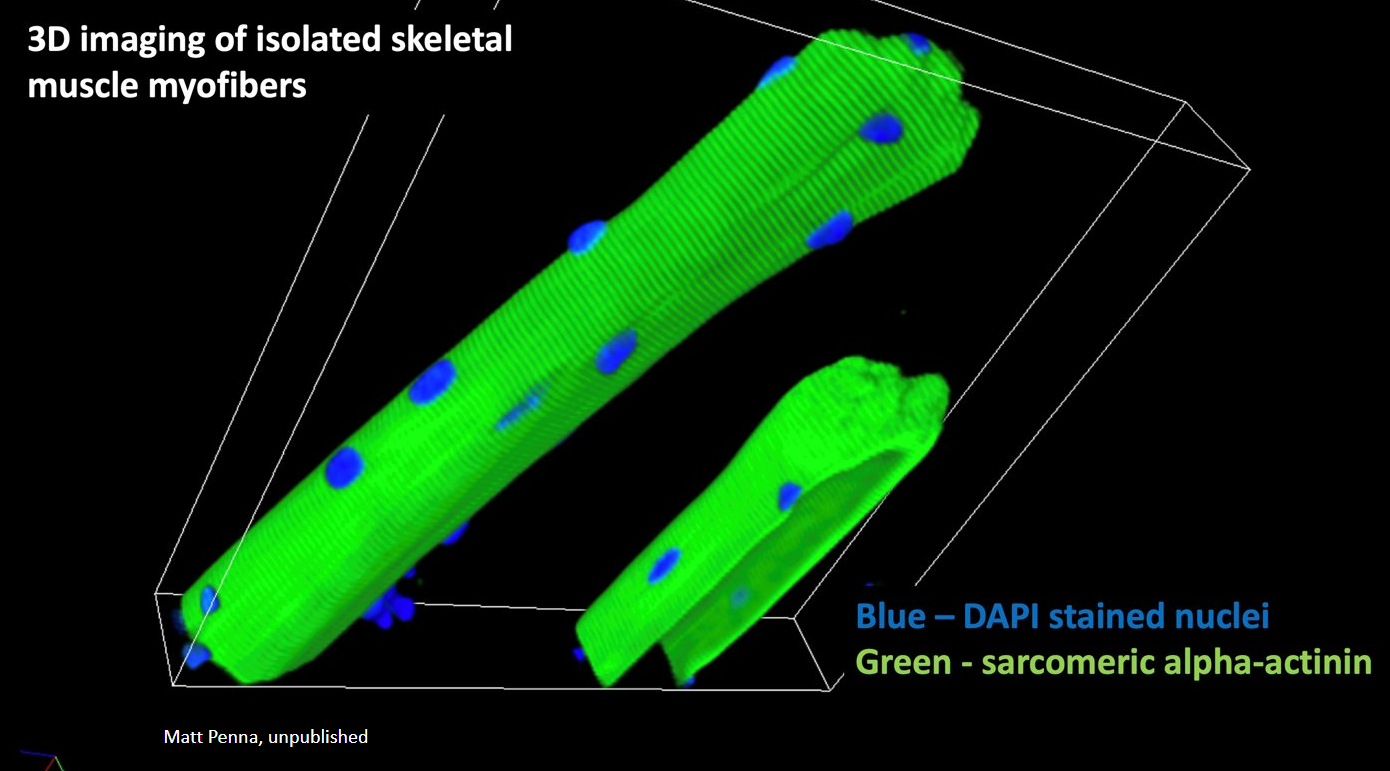
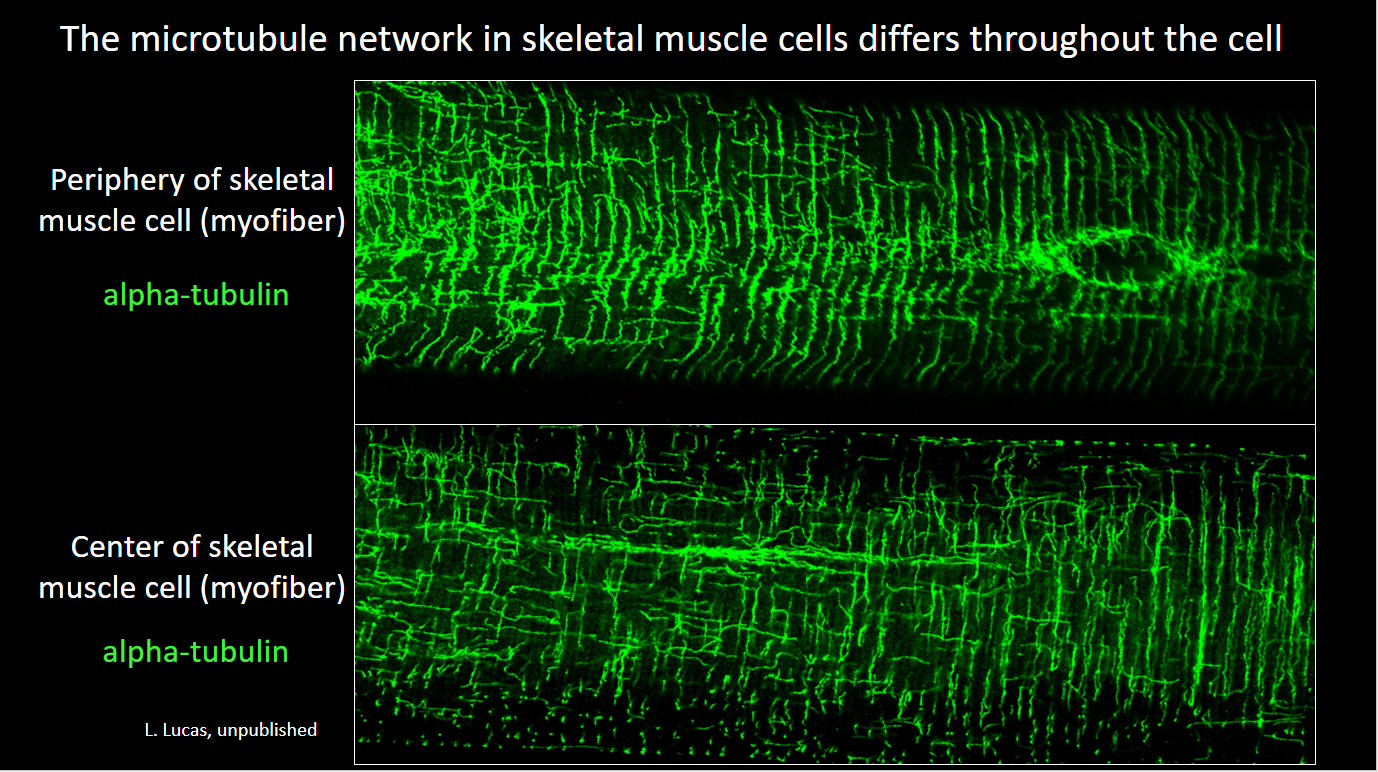
Lathan’s paper on the role of the striated muscle splice variant of Map4 in skeletal muscle microtubule structure was published in iScience, 2024
Ashwini joined our lab as a new Ph.D. student, 2024
Ashwini is an awardee of the Trentin Award for academic achievement 2024
Sara’s paper that identifies HSP90 as a modifier of RNA foci in myotonic dystrophy type was published in Mol. Cell Biol. 2024
Sara defended her thesis and graduated 2024
Larissa’s paper demonstrating the reduced expression of Scn5a in myotonic dystrophy cardiac tissues was published in Hum. Mol. Genet., 2024
Andrew received the BioSciences Award for Outstanding Research voted by the faculty of the Department of BioSciences at Rice University. May 2024.
Janel won best oral presentation for basic research at the biannual International Myotonic Dystrophy Consortium Meeting in Nijmegen, Netherlands April 2024.
Larissa’s perspective of a paper from the John Lueck lab was published in J. Clinical Investigation
Larissa was awarded a three year NIH F32 postdoctoral fellowship award to start January 2024
Reagents
The following plasmids are available through Addgene.
FRE5 (FLAGNLSREDGFP5)
FRE5Cf (FLAGNLSREDGFP5Claf)
RG6
RHCglo
RTB300
R300TA
DT960
DT480
DT240
DT40
DT12A
DT0
pBItetDT960GFP
pBItetDT12nGFP
FLYLQ (Flag-CELF1)
pBItetDMPKSGFP
pBItetDT240GFP
pBItetDT480GFP
FlagETR3 (CELF2)
FlagMBNL1-41
FlagMBNL3
Alternative splicing is crucial to muscle mass maintenance
Alternative splicing allows cells to make many different proteins with a limited number of genes.
There is more going on in DM1 than just alternative splicing
In his laboratory at Baylor College of Medicine, Dr. Thomas A. Cooper is leading the way to better understand myotonic dystrophy type 1 (DM1), a rare but devastating condition.
What causes myotonic dystrophy, type1
The lab of Dr. Thomas Cooper discovered that the CUGexp RNA disrupts the regulation of RNA splicing that is important for proper functions of a large number of genes.
Image of the Month: From the Labs blog









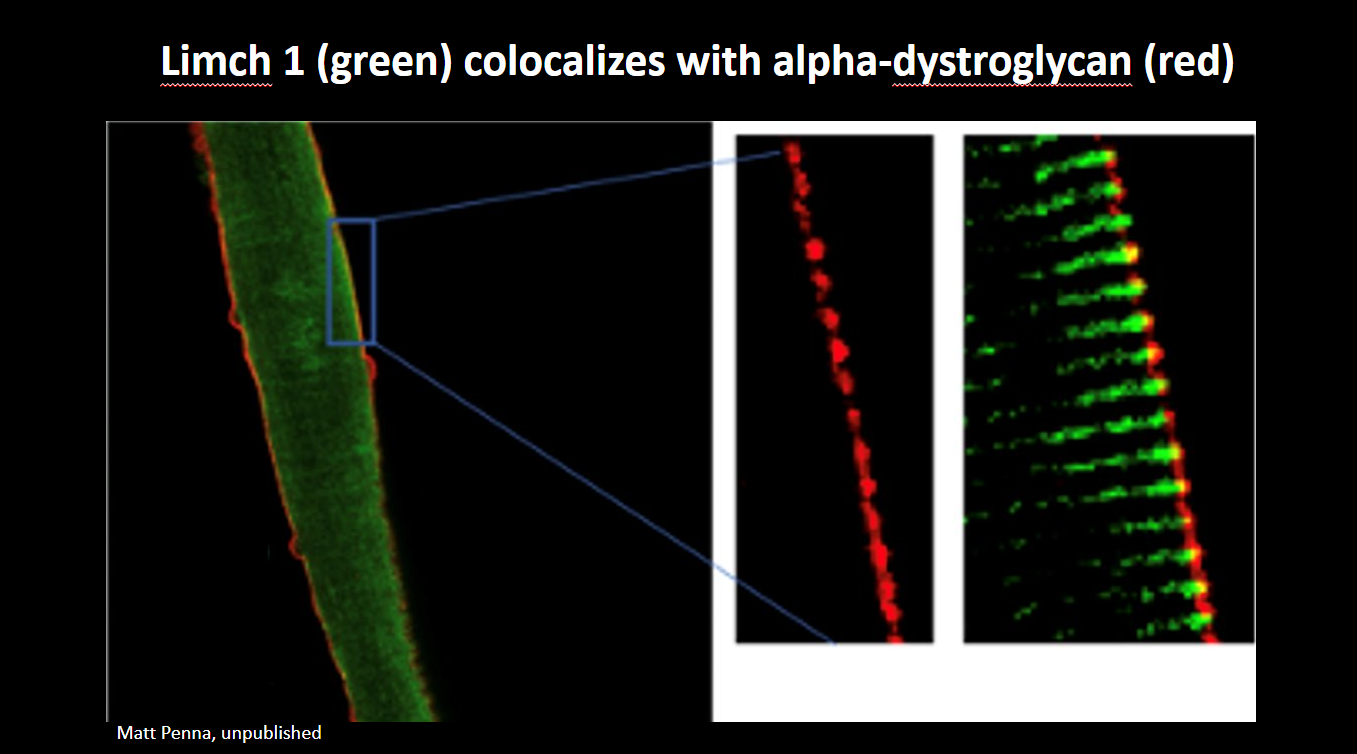
 Credit
Credit