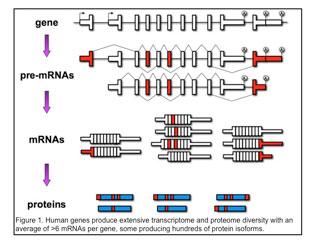
Individual genes express multiple mRNAs by pre-mRNA alternative splicing, alternative polyadenylation or use of alternative promoters (first exons). As a result, individual genes express multiple mRNAs that are identical except for discrete regions of variability (indicated in red in Figure 1)
More than 95 percent of human genes undergo alternative splicing and >50 percent of genes use alternative polyadenylation sites. For many genes, pre-mRNA processing directs expression of functionally divergent protein isoforms according to cell-specific regulation based on differentiated cell type, developmental stage, gender, or in response to external signals. Alternative splicing and alternative polyadenylation also generates variability within mRNA untranslated regions and affects gene expression by adding or removing cis-acting elements that regulate mRNA translation efficiency, mRNA stability, or mRNA intracellular localization. In addition, up to 30 percent of alternative splicing changes introduce a premature termination codon that lead to mRNA down-regulation by nonsense mediated decay such that alternative splicing can secondarily regulate mRNA abundance.
Technological and computational approaches have made spectacular advances allowing genome-wide identification of transcriptome changes (RNA-seq), translation (ribosome profiling), binding sites of RNA binding proteins in vivo (CLIP-seq), RNA secondary structure, and sites of RNA methylation, to mention a few. It is now clear that regulation of alternative splicing has a huge impact on gene function and there is a great deal to learn about questions ranging from the mechanisms of coordinated alternative splicing to the functional consequences of the divergent isoforms expressed from individual genes. We have identified robust networks of coordinated regulation driven by a number of RNA binding proteins that make substantial contributions to the physiological transition from fetal to adult tissue function and are required for normal function and maintenance of adult tissues (see Projects Mechanisms and Consequences of Coordinated Splicing Regulation).
Splicing and its disruption have a major role in causing disease as well as in modifying disease severity. We study the pathogenic mechanisms as well as therapeutic approaches for myotonic dystrophy, in which a microsatellite expansion in the DMPK gene has a trans-dominant effect on regulation of alternatively spliced pre-mRNAs causing major features of the disease (see Projects Mechanism of Myotonic Dystrophy Pathogenesis) The effect is to reverse developmentally regulated splicing of a subset of genes such that fetal splicing patterns are expressed in adult tissues. This results in features of the disease since fetal isoforms cannot perform functions that are required in adult tissues.
References
Braunschweig, U., Gueroussov, S., Plocik, A. M., Graveley, B. R. and Blencowe, B. J. Dynamic Integration of Splicing within Gene Regulatory Pathways. Cell 152, 1252–1269 (2013).
Tian, B. and Manley, J. L. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 38, 312–320 (2013).
Kalsotra, A. and Cooper, T. A. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 12, 715–729 (2011).
Ellis, J. D. et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol. Cell 46, 884–892 (2012).
Buljan, M. et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell 46, 871–883 (2012).
Acuña, L. I. G. and Kornblihtt, A. R. Long range chromatin organization. Transcription 5, e28726 (2014).
Wang, G.-S. and Cooper, T. A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 8, 749–761 (2007).
Cooper, T. A., Wan, L. and Dreyfuss, G. RNA and disease. Cell 136, 777–793 (2009).








