The lab has a long-standing interest in the use of molecular libraries to answer questions relevant to enzyme structure and function, discovery of protein-protein interaction inhibitors, and examination of the human humoral immune response to viral infections.
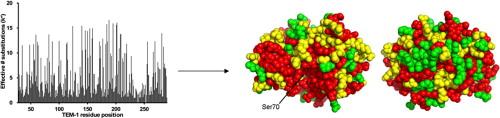
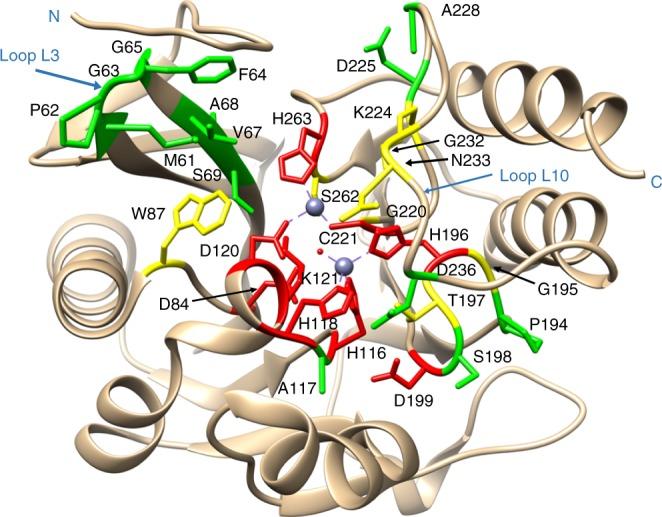
We have used codon randomization followed by functional selections and deep sequencing to study the determinants of enzyme catalysis and substrate specificity with a focus on β-lactamases.
Deng, Z., Huang, W., Bakkalbasi, E., Brown, N.G., Adamski, C.J., Rice, K., Muzny, D., Gibbs, R.A., and Palzkill, T. (2012). Deep sequencing of systematic combinatorial libraries reveals -lactamase sequence constraints at high resolution. J. Mol. Biol. 424: 150-167.
Sun, Z., Hu, L., Sankaran, B., Prasad, B.V.V., and Palzkill, T. (2018). Differential active site sequence requirements for NDM-1 β-lactamase hydrolysis of carbapenem versus penicillin and cephalosporin antibiotics. Nat. Commun. 9:4524
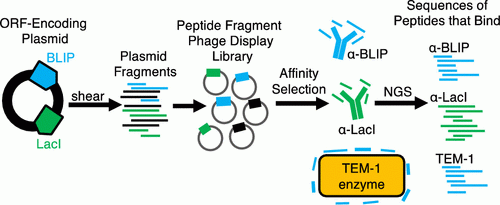
We are using phage display methods coupled with deep sequencing to map peptides originating from protein-protein interaction interfaces that retain binding affinity for the partner protein. We have shown that these peptides, when synthesized can disrupt protein-protein interactions and can serve as starting points for the development of therapeutics that modulate protein-protein interactions.
Huang, W., Soeung, V., Boragine, D.M., and Palzkill, T. (2020). Mapping protein-protein interaction interface peptides with Jun-Fos assisted phage display and deep sequencing. ACS Synth. Biol. 9:1882-1896.
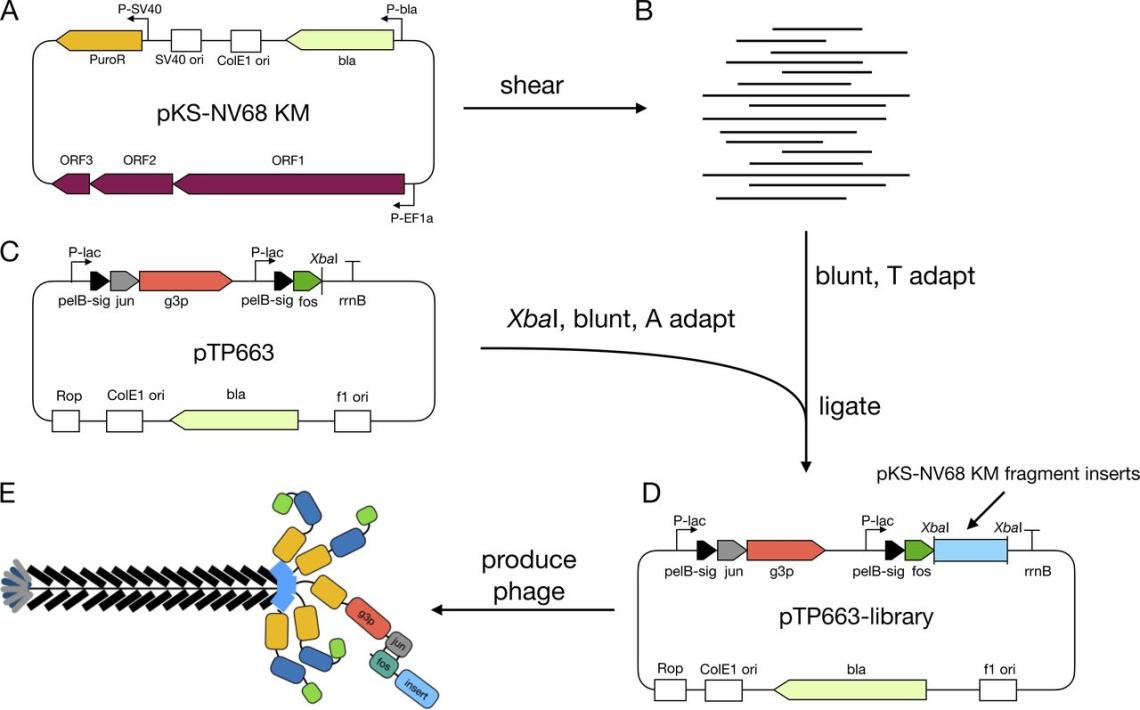
We have developed methods to construct genomic phage display libraries that can be used with complex samples such as patient serum samples and coupled with deep sequencing to map multiple epitopes simultaneously and thus determine the antigenic landscape of the humoral immune response to infections. We recently demonstrated this approach with a Norovirus genomic phage display library and immune serum to map epitopes recognized by the humoral immune response to Norovirus infection.
Huang, W., Soeung, V., Boragine, D.M., Hu, L., Prasad, B.V.V., Estes, M.K., Atmar, R.L., and Palzkill, T. (2020). High-resolution mapping of human norovirus antigens via genomic phage display library selections and deep sequencing. J. Virol. 95:e01495-20.