We are interested in how sequence determines function in enzymes. In particular, we investigate the structural and mechanistic basis for antibiotic resistance enzyme function. In this regard, we are using pre-steady state, stopped-flow kinetics experiments to determine the rate-limiting steps for hydrolysis of β-lactam antibiotics by important antibiotic resistance enzymes including the clinically widespread KPC and CTX-M β-lactamases. These enzymes are encoded on plasmids and spread rapidly among Gram-negative bacteria by gene transfer. Importantly, these enzymes catalysis the hydrolysis of clinically-relevant β-lactam antibiotics to cause drug resistance and thereby limit treatment options. We couple the study of enzyme kinetics with X-ray crystal structure determinations to link structure with mechanism. We have determined high resolution structures of wild-type enzymes as well as drug-resistant variant enzymes that have evolved due to the selective pressure of antibiotic therapy. The results of these studies will guide the rational design of antibiotics that resist the action of β-lactamases.
We are also interested in the structure and mechanism of the Mobile Colistin Resistance (MCR) enzyme that provides Gram-negative bacteria with resistance to polymyxin antibiotics such as colistin. MCR resides in the bacterial cytoplasmic membrane and transfers phosphoethanolamine from phosphatidylethanolamine to lipid A. This reaction results in neutralization of the negative charge on the surface of bacteria, which blocks the binding and action of colistin. We determined the first structure of the soluble, catalytic domain of MCR-1 and are currently working on the structure and mechanism of the full-length enzyme.
Brown, C.A., Hu, L., Sun, Z., Patel, M.P., Singh, S., Porter, J.R., Sankaran, B., Prasad, B.V.V., Bowman, G.R., and Palzkill, T. (2020). Antagonism between substitutions in β-lactamase explains a path not taken in the evolution of bacterial drug resistance. J. Biol. Chem. 295:7376-7390.
Mehta, S.C., Furey, I.M., Pemberton, O.A., Boragine, D.M., Chen, Y., and Palzkill, T. (2021). KPC-2 β-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate. J. Biol. Chem. 296:100155.
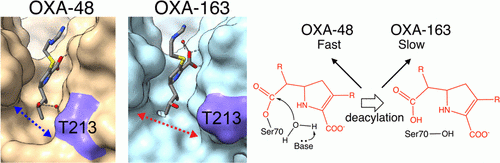
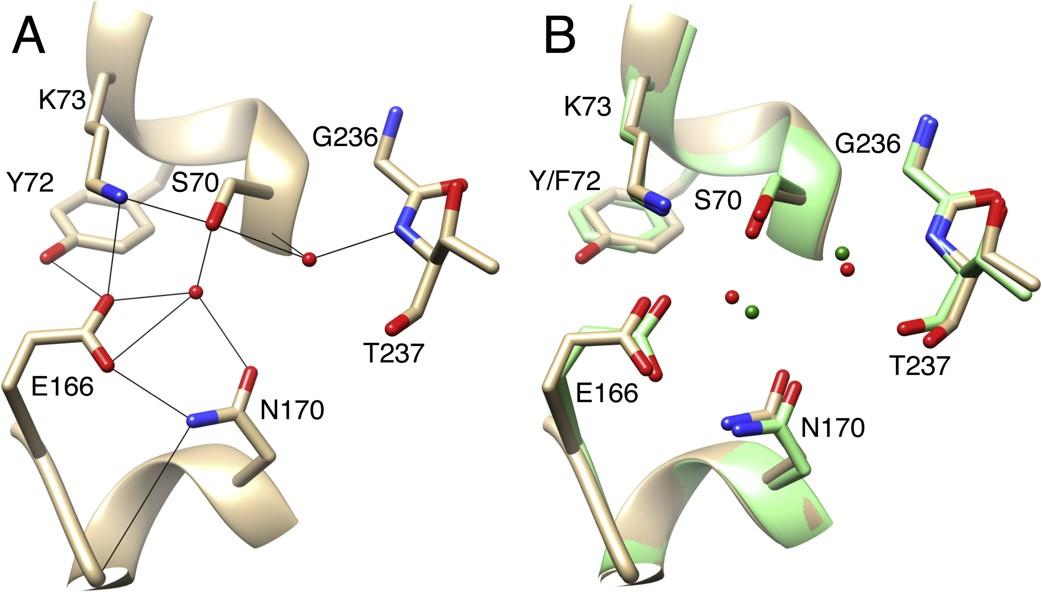
Stojanoski, V., Hu, L., Sankaran, B., Wang, F., Tao, P., Prasad, B.V.V., and Palzkill, T. (2021). Mechanistic basis of OXA-48 β-lactamases’ hydrolysis of carbapenems. ACS Infect. Dis. 7:445-460.
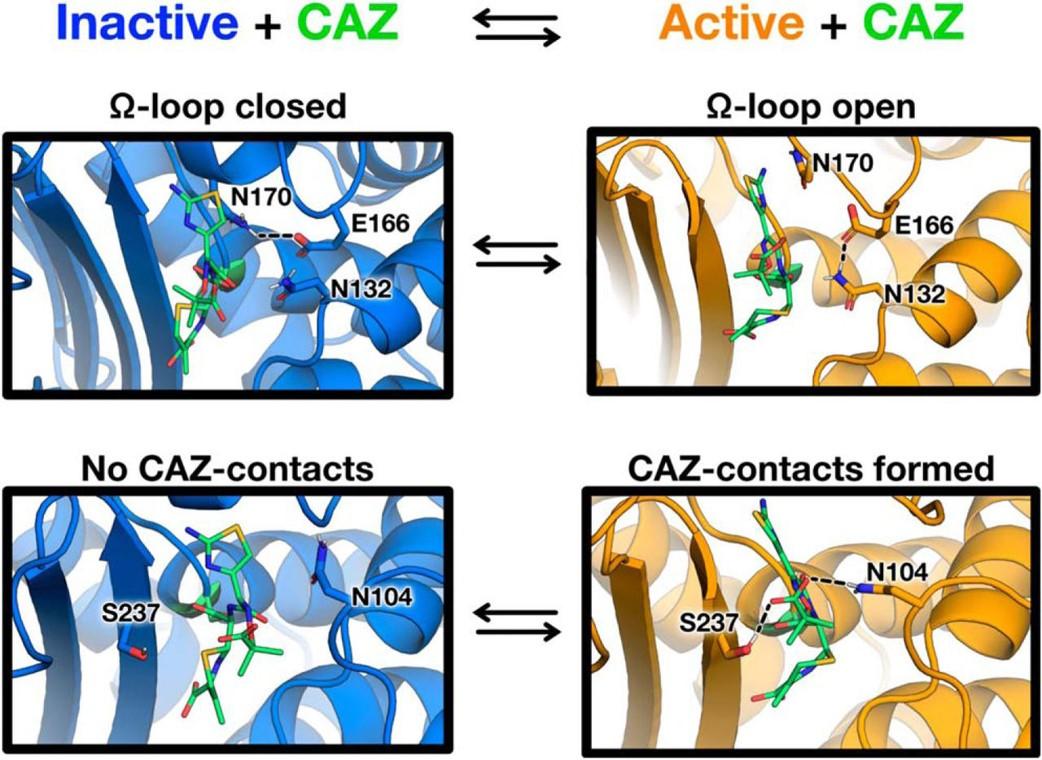
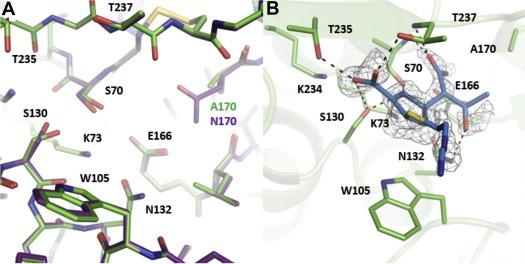
Furey, I.M., Mehta, S.C., Sankaran, B., Hu, L., Prasad, B.V.V., and Palzkill, T. (2021). Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 β-lactamase. J Biol Chem. 296:100799.