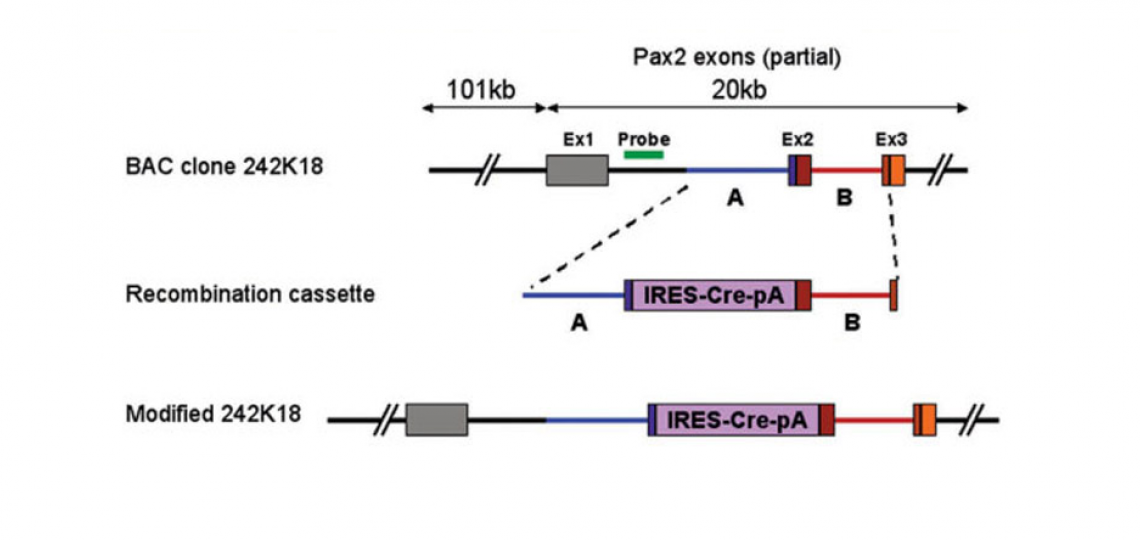
Pax2-Cre Transgenic Mice
These mice were generated in our lab at the House Ear Institute by Taka Ohyama. The Cre recombinase gene was inserted into a bacterial artificial chromosome (BAC) containing about 120kb of the Pax2 genomic locus by homologous recombination in bacteria.
The Genesis paper describing the construction and characterization of these mice is available.
Pax2-Cre mice are available for purchase from the Mutant Mouse Regional Resource Center in North Carolina.
A generic Cre genotyping protocol for the mice is available. A more specific genotyping protocol that identifies the IRES-Cre element specifically is also available, but bear in mind that this protocol will detect other IRES-Cre transgenes too.
We and others have observed that the Pax2-Cre transgene can be active in the female germline. There have been isolated reports of Pax2 being expressed in female gonads, which may explain this result. Using Pax2-Cre females to create conditional mutants can therefore give embryos with recombination throughout the body. This effect seems to have variable penetrance depending on the background strains of the mice and the floxed allele involved. For this reason, we recommend that only Pax2-Cre males are used to generate conditional mutants
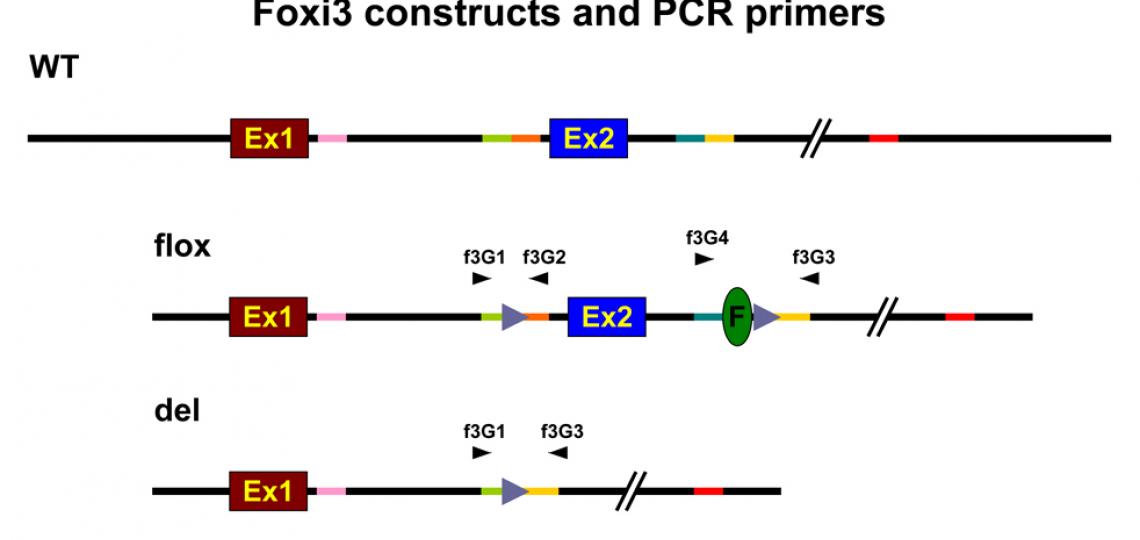
Foxi3 Mutant Mice
Foxi3 mutant mice were generated in our lab at the House Ear Institute by Taka Ohyama. The loxP-flanked allele can be turned into a null allele by mating with Cre-expressing lines, which removes exon 2.
Foxi3-flox mice are available for purchase from the Jackson Laboratory.
View a genotyping protocol for the mice.
Shirokova, V., Jussila, M., Biggs, L.C., Ohyama, T., Groves, A.K. and Mikkola, M.L. (2016). Foxi3 deficiency compromises hair follicle stem cell specification and activation. Stem Cells 10.1002/stem.2363
Birol, O., Ohyama, T., Edlund, R.K., Drakou, K., Georgiades, P., and Groves, A.K. (2015). The mouse Foxi3 transcription factor is necessary for the development of posterior placodes. Developmental Biology 409, 139-151.
Jussila, M., Aalto, A., Sanz Navarro, M., Shirokova, V., Kallonen, A., Ohyama, T., Groves, A.K., Mikkola, M.L. and Thesleff, I. (2015). Suppression of epithelial differentiation by Foxi3 is essential for molar crown patterning. Development 142, 3954-3963.
Edlund, R.K., Ohyama, T., Kantarci, H., Riley, B.B. and Groves, A.K. (2014). Foxi transcription factors promote pharyngeal arch development by regulating the formation of FGF signaling centers. Developmental Biology 390, 1-13.
Foxi3-GFP and Foxi3-CreER Mice
Foxi3-GFP and Foxi3-CreER mice were generated at Baylor College of Medicine by Andy Groves and Onur Birol. For Foxi3-GFP mice, we targeted the Foxi3 locus to insert a GSG-P2A-Venus fluorescent protein sequence downstream of the Foxi3 coding region. For Foxi3-CreER mice, we replaced both Foxi3 coding exons and the intervening intron with a construct containing CreER, a GSG-P2A sequence followed by EGFP. We found that although the Foxi3-CreER mice express CreER efficiently, we were not able to detect EGFP in these mice. The Foxi3-CreER allele acts as a functional null allele.
Foxi3-GFP mice are available for purchase from the Jackson Laboratory. View genotyping information.
Foxi3-CreER mice are available for purchase from the Jackson Laboratory. View genotyping information.
Thawani, A., Maunsell, H.R., Zhang, H., Ankamreddy, H. and Groves, A.K. (2023). The Foxi3 transcription factor is necessary for the fate restriction of placodal lineages at the neural plate border. Development 150, dev202047.
Ankamreddy, H., Thawani, A., Birol, O., Zhang, H., and Groves, A.K. (2023). Foxi3GFP and Foxi3CreER mice allow identification and lineage labeling of pharyngeal arch ectoderm and endoderm, and tooth and hair placodes. Developmental Dynamics 252,1462-1470. doi: 10.1002/dvdy.645
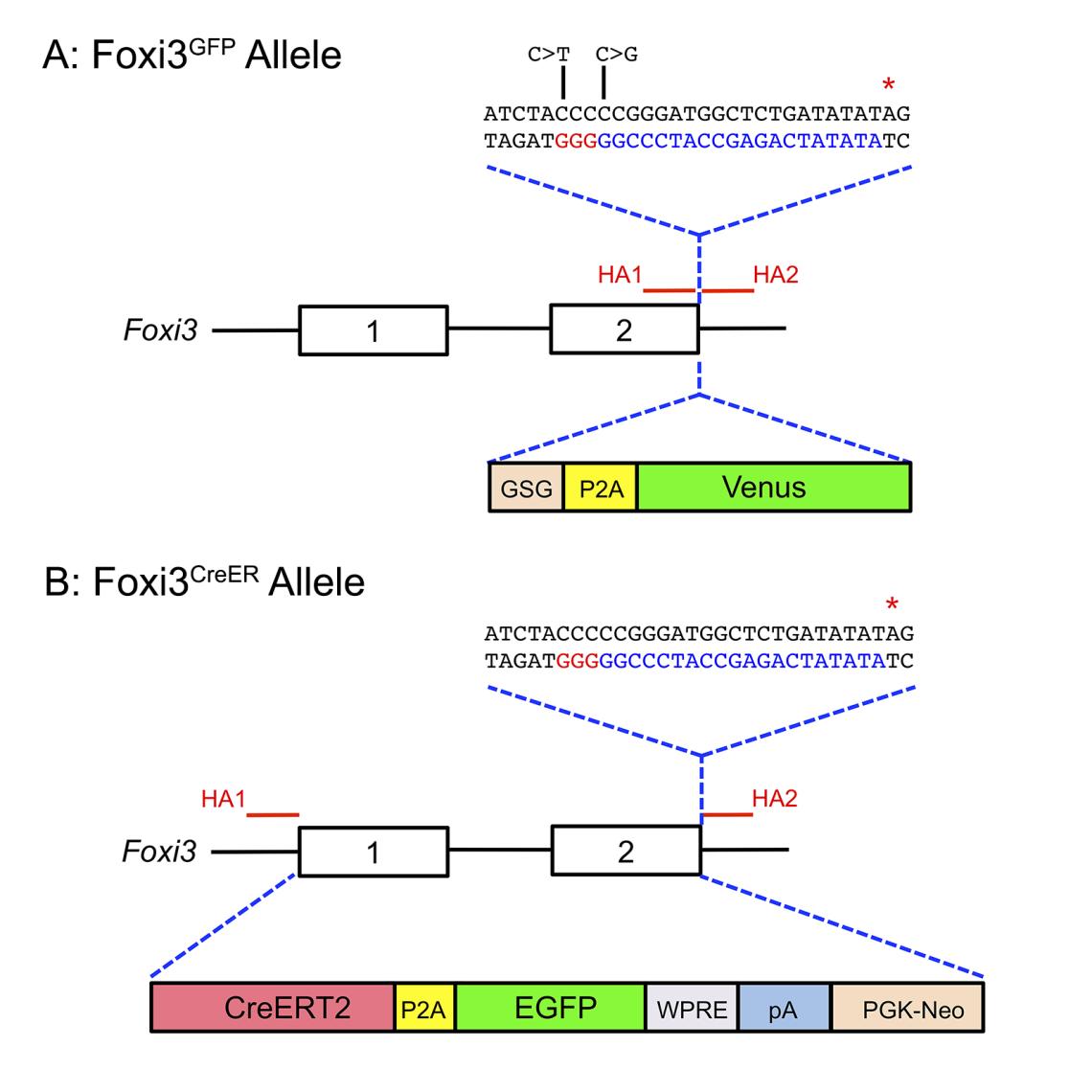
A) Diagram showing the Foxi3 locus, including the position of the two homology arms (HA1 and HA2) used to target the locus by CRISPR-mediated homology-directed repair. The homology arms enclosed a DNA sequence coding for a GSG-P2A-Venus fluorescent protein, followed by a terminating stop codon (red asterisk). The sequence corresponding to the gRNA targeting the 3’ end of Foxi3 is shown in blue letters, with the PAM sequence shown in red letters. The DNA used for homologous recombination contained two nucleotide changes (C>T and C>G; indicated in the diagram) to prevent the inserted sequence from being targeted by the Foxi3 gRNA.
(B) Diagram showing the Foxi3 locus, including the position of the two homology arms (HA1 and HA2) used to target either end of the Foxi3 gene and insert CreER by homologous recombination in ES cells. Exons 1 and 2 and the intervening intron were replaced by an insert containing a CreERT2 fusion cDNA, a P2A sequence followed by EGFP, a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), a polyA signal, and a PGK-neo resistance cassette. Homologous recombination in ES cells was enhanced by CRISPR-mediated cleavage of the 3’ end of the Foxi3 locus using the same gRNA as in (A) above.








