About the Unit
The diagnostic challenges posed by undiagnosed diseases have solutions in animal models that precisely model human disease-associated genetic variation. Genetically modified Drosophila melanogaster (fly) and Mus musculus (mouse), are critical tools for interpretation of variants identified in human through exome and genome sequencing. Flies are an effective model organism to rapidly characterize the functionally consequence of disease-associated variants and to further probe pathogenic mechanisms. Mice are essential for mimicking human disease phenotype, modeling variants that affect systems for which fly may not be appropriate, and for pre-clinical therapeutic studies. Moreover, naturally occurring, spontaneous mutations remain an important resource for human disease modeling, particularly, in non-human primates. In non-human primates, these types of mutations can provide important models for coronary artery disease, social cognition, addiction, depression, and autism.
The Disease Modeling Unit is responsible for producing and phenotyping the center’s precision fly and mouse models. The unit leverages the genome modification techniques and phenotyping capabilities of the Undiagnosed Diseases Network Model Organism Screening Center (UDN MOSC) Fly Core and the Knockout Mouse Phenotyping Project (KOMP2) site at Baylor. Moreover, the unit uses a resource of over 800 rhesus macaque whole genome and exome sequences with paired phenotype information from the Human Genome Sequencing Center (HGSC) at Baylor to identify putative disease models in the national primate colonies. This provides investigators who are interested in a primate disease model with initial phenotype-genotype information and a potential collaborator for additional primate studies.
Submitted Variant Review and Model Selection
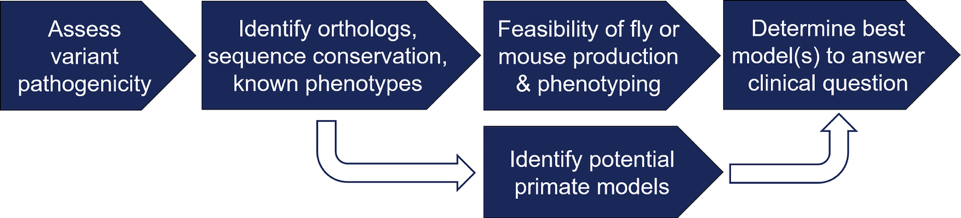
During the center’s review and selection of submitted variants for modeling, the unit plays a critical role in determining the feasibility of developing a fly or mouse model and determining the availability of phenotyping strategies that can answer clinically relevant questions.
Model Development and Characterization
Once selected for modeling by the center and an experimental plan is established, the Unit uses advanced genome modification techniques to produce mouse or fly models that recapitulate the human disease-associated variant and phenotyping workflows that validate variant-disease associations, identify pathogenic mechanisms, and test putative interventions. If a potentially informative primate genetic variant is identified, the unit works with our primate center collaborators to identify specific animals that may be useful for inferring genotype-phenotype associations.
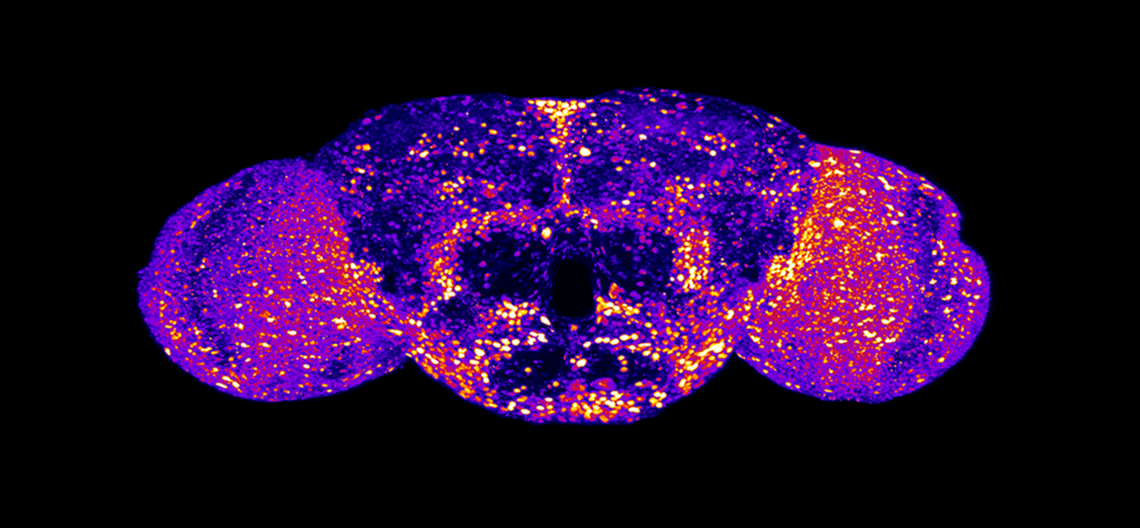
Shown above: Precision fruit fly (Drosophila) models, in which expression of the endogenous fly Uba5 gene is replaced by human UBA5 cDNA, classified 12 developmental and epileptic encephalopathy 44 (DEE44)-associated variants as causing mild, intermediate, or severe loss of function. This is a representative image of endogenous Uba5 expression levels in the adult fly brain. Data were transformed into a heatmap image. Bright, warm colors (red and yellow) indicates high expression levels; dark, cold colors (blue and purple) indicates lower expression levels. See Pan et al., PMID: 38079206.
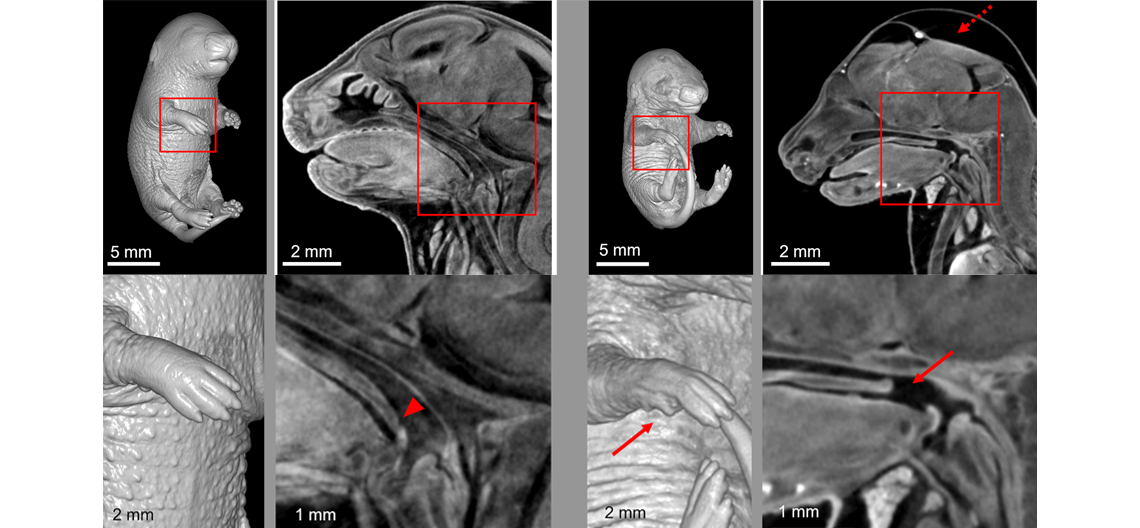
Shown above: A precision mouse model verified the pathogenicity of a p.E66K variant in AXIN2 found in individuals with phenotypic expansion of AXIN2-related disorders. Heterozygosity for the p.E66K variant is perinatal lethal and causes shortened soft palate and skeletal abnormalities in mouse. Left Images: Representative micro-computed tomography imaging of a wild-type E18.5 embryo. No external anomalies are present, and the soft palate is the expected length. Right Images: Representative micro-computed tomography imaging of an Axin2 p.E66K expressing embryo with a missing forelimb digit, edema, and a shortened soft palate. Bottom images: magnified views of the areas in red boxes on the top images. Dashed arrow indicates edema; arrowhead indicates normal soft palate; arrows indicate a missing digit and a shortened soft palate. See Aceves-Ewing et al., PMID: 39677486.
Core Members
- Jason Heaney, Ph.D. (Lead)
- Shinya Yamamoto, D.V.M., Ph.D. (Co-Lead)
- Jeffrey Rogers, Ph.D.
- Michael Wangler, M.D.
- Chih-Wei Logan Hsu, Ph.D.
- Audrey Christiansen, Ph.D.








 Credit
Credit