About the Lab
Our research area is the study of molecular mechanisms of retinal development.
The primary goal of our research is to understand the molecular mechanisms of retinal development with the ultimate goal of improving our ability prevent, diagnose, and treat human retinal disease. To this end, we are using two animals, the mouse Mus musculus and the fruit fly Drosophila melanogaster, as model systems to identify and determine the function of conserved genes required for normal retinal development. In spite of substantial differences between vertebrate and insect retinal morphology, genetic mechanisms of retinal development have been conserved for more than 500 million years. Thus, study of the molecular and genetic pathways controlling Drosophila eye development has provided a valuable set of tools with which to decipher the development and function of the vertebrate retina. In addition, the mouse provides a powerful model system to decipher the function of newly identified human retinal disease genes and to conduct gene therapy studies.
Fruit Fly Drosophila Melanogaster
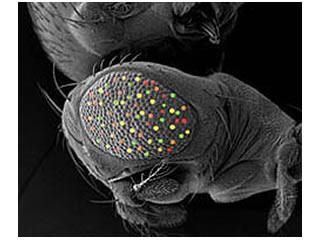
Our main Drosophila project uses a combinatorial approach of genetics, genomics, and computational biology to dissect the roles of four retinal determination genes: eyeless, toy, eyes absent, and sine oculis, all of which encode highly conserved transcription factors that are both necessary and sufficient for eye development.
We are using molecular genetics and genomics to identify direct targets of these transcription factors and the pathways they regulate to control normal retinal development. In addition, we are using novel genomic rescue strategies to definitively dissect the in vivo function of these genes.
Finally, we are using the CRISPR/Cas9 system to create new alleles to conduct functional studies of eyeless and toy.
Mouse Mus Musculus
In the mouse, we have created knockout models for two genes that cause congenital blindness in humans, Spata7 and Kcnj13. Loss-of-function of these genes results in early visual system defects in both humans and in our mouse models, which fully recapitulate the human disease condition. We are using these models to fully dissect the molecular mechanisms by which the genes act in vivo and as tools for conducting gene therapy studies.
Our most recent models for Kcnj13 have been created using CRISPR/Cas9 and have greatly increased the speed with which we can dissect gene function in vivo.








