Herman Lab research can be most easily divided into three major domains, with each member of the lab more or less specializing in one for their project:
- Genome Stability
- Transcription Infidelity
- Horizontal Gene Transfer
Genome Stability
It’s known that the genome stability of a cell is a major determinant of its fitness, as genomic strand breaks, re-arrangements, or single nucleotide mutations stand to compromise proper cell division, global transcriptional programs, and the expression of essential genes. It is also known that proper transcription of the same genome is a determinant of cellular fitness as mis-transcribed or partially transcribed mRNA’s lead to many defective proteins that are toxic to the cell. However, both of these processes, DNA replication and RNA transcription, occur on the same template DNA. As such, they are bound to conflict.
These conflicts are one of our lab’s subjects of interest. We seek to understand how these replication-transcription conflicts occur, how they are resolved, and what accessory factors mediate these conflicts.
Much of our work, in this regard has brought our focus to RNA polymerase (RNAP) secondary channel interactors. These factors bind in the RNAP secondary channel, which is thought to primarily function as a intake funnel for dNTPs, and modulate RNAP structure, function, and attributes. Our model, Escherichia coli Str. K12, has three such factors: greA, greB, and dksA.
Our group and our collaborators have made major breakthroughs in understanding how these secondary channel factors interact with DNA replication and DNA repair; to assist or conflict with the stability of the genome.
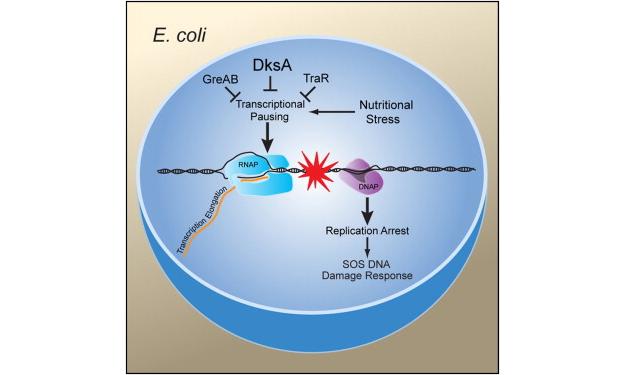
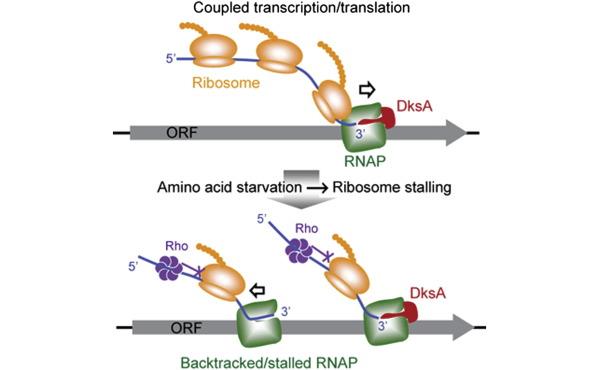
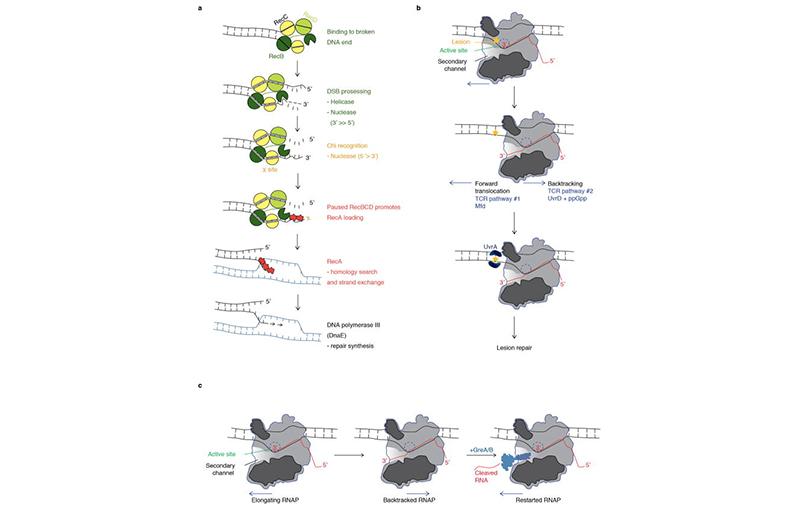
For more information on this exciting research, please refer to the following publications:
Yin Zhai, P.J. Minnick, John P. Pribis, Libertad Garcia-Villada, P.J. Hastings, Christophe Herman, Susan M. Rosenberg. ppGpp and RNA-Polymerase Backtracking Guide Antibiotic-Induced Mutable Gambler Cells. bioRxiv 2022.07.06.499071; doi: https://doi.org/10.1101/2022.07.06.499071.
Priya Sivaramakrishnan, Catherine C. Bradley, Irina Artsimovitch, Katelin M. Hagstrom, Laura Deus Ramirez, Alice Xueyin Wen, Matthew Brandon Cooke, Maya Shaulsky, Christophe Herman, Jennifer A. Halliday. Modulation of RNA polymerase processivity affects double-strand break repair in the presence of a DNA end-binding protein. bioRxiv 2022.02.08.479637; doi: https://doi.org/10.1101/2022.02.08.479637.
Xia J, Chiu LY, Nehring RB, Bravo Núñez MA, Mei Q, Perez M, Zhai Y, Fitzgerald DM, Pribis JP, Wang Y, Hu CW, Powell RT, LaBonte SA, Jalali A, Matadamas Guzmán ML, Lentzsch AM, Szafran AT, Joshi MC, Richters M, Gibson JL, Frisch RL, Hastings PJ, Bates D, Queitsch C, Hilsenbeck SG, Coarfa C, Hu JC, Siegele DA, Scott KL, Liang H, Mancini MA, Herman C, Miller KM, Rosenberg SM. Bacteria-to-Human Protein Networks Reveal Origins of Endogenous DNA Damage. Cell. 2019 Jan 10;176(1-2):127-143.e24. doi: 10.1016/j.cell.2018.12.008. PMID: 30633903; PMCID: PMC6344048.
Sivaramakrishnan P, Sepúlveda LA, Halliday JA, Liu J, Núñez MAB, Golding I, Rosenberg SM, Herman C. The transcription fidelity factor GreA impedes DNA break repair. Nature. 2017 Oct 12;550(7675):214-218. doi: 10.1038/nature23907. Epub 2017 Oct 4. PMID: 28976965; PMCID: PMC5654330.
Kotlajich MV, Xia J, Zhai Y, Lin HY, Bradley CC, Shen X, Mei Q, Wang AZ, Lynn EJ, Shee C, Chen LT, Li L, Miller KM, Herman C, Hastings PJ, Rosenberg SM. Fluorescent fusions of the N protein of phage Mu label DNA damage in living cells. DNA Repair (Amst). 2018 Dec;72:86-92. doi: 10.1016/j.dnarep.2018.09.005. Epub 2018 Sep 14. PMID: 30268364; PMCID: PMC6287932.
Xia J, Chen LT, Mei Q, Ma CH, Halliday JA, Lin HY, Magnan D, Pribis JP, Fitzgerald DM, Hamilton HM, Richters M, Nehring RB, Shen X, Li L, Bates D, Hastings PJ, Herman C, Jayaram M, Rosenberg SM. Holliday junction trap shows how cells use recombination and a junction-guardian role of RecQ helicase. Sci Adv. 2016 Nov 18;2(11):e1601605. doi: 10.1126/sciadv.1601605. PMID: 28090586; PMCID: PMC5222578.
Zhang Y, Mooney RA, Grass JA, Sivaramakrishnan P, Herman C, Landick R, Wang JD. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol Cell. 2014 Mar 6;53(5):766-78. doi: 10.1016/j.molcel.2014.02.005. PMID: 24606919; PMCID: PMC4023959.
Shee C, Cox BD, Gu F, Luengas EM, Joshi MC, Chiu LY, Magnan D, Halliday JA, Frisch RL, Gibson JL, Nehring RB, Do HG, Hernandez M, Li L, Herman C, Hastings PJ, Bates D, Harris RS, Miller KM, Rosenberg SM. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. Elife. 2013 Oct 29;2:e01222. doi: 10.7554/eLife.01222. PMID: 24171103; PMCID: PMC3809393.
Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010 May 14;141(4):595-605. doi: 10.1016/j.cell.2010.03.036. PMID: 20478253; PMCID: PMC2919171.
Transcription Infidelity
Much of modern molecular genetics has been concerned with the concept of error propagation in DNA. In basic research, we refer to these as mutations – our more clinically inclined friends: variants. Semantics aside, these mutations are the basis of long term cellular adaptation; they are the only event that can heritably change a cell’s phenotype and fitness – or are they?
Our lab aims to adjust this notion and make a case that errors in transcriptional RNA propagation are also able to influence cellular phenotypes, and subsequently, fitness.
In Escherichia coli many genes rely on a relative sparsity of mRNA, with anywhere from 1-5 transcripts per cell cycle. These mRNA’s are then used to translate orders of magnitude more protein. Finally, these proteins are generally quite stable – an average prokaryotic protein has a half life of 20 hours, whereas the cell doubling time can be lower than 0.25 hours. With these facts in mind, a single error on a single mRNA can impact the end-point activity or expression of a gene for multiple generations. This transient, but heritable, phenotype, at the population level, may provide a fitness benefit in changing conditions. Since the rate of RNA errors is roughly 1,000 to 10,000 fold higher than the rate or DNA errors, this transient fitness phenotype may play a larger role than previously thought.
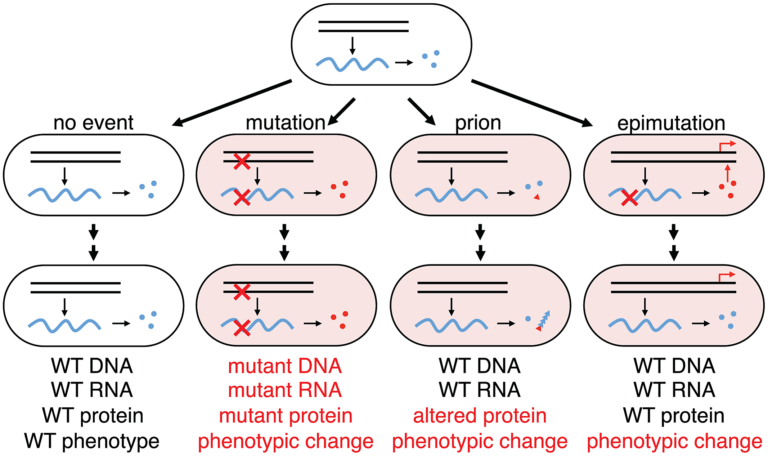
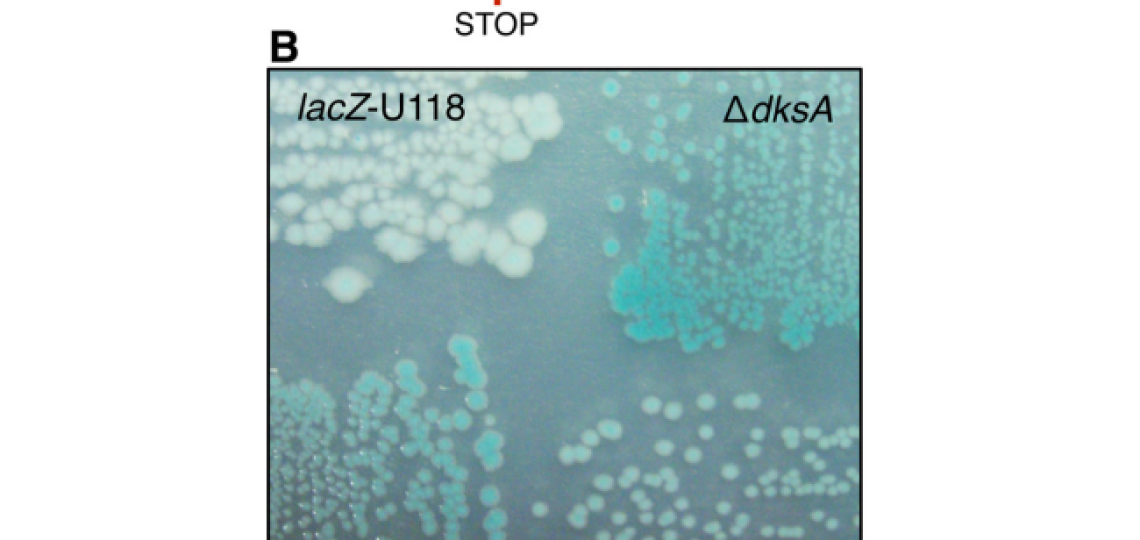
We aim to expand upon this research by identifying additional determinants of transcriptional errors and further investigating their single cell and population effects. For more reading on this subject, refer to the following publications:
Christophe Herman, Catherine Bradley, Alasdair Gordon, Chen Wang, Matthew Cooke, Brendan Kohrn, Scott Kennedy, Olivier Lichtarge, Shannon Ronca. RNA polymerase inaccuracy underlies SARS-CoV-2 variants and vaccine heterogeneity.
Bradley CC, Gordon AJE, Halliday JA, Herman C. Transcription fidelity: New paradigms in epigenetic inheritance, genome instability and disease. DNA Repair (Amst). 2019 Sep;81:102652. doi: 10.1016/j.dnarep.2019.102652. Epub 2019 Jul 8. PMID: 31326363; PMCID: PMC6764924.
Gordon AJE, Sivaramakrishnan P, Halliday JA, Herman C. Transcription infidelity and genome integrity: the parallax view. Transcription. 2018;9(5):315-320. doi: 10.1080/21541264.2018.1491251. Epub 2018 Aug 10. PMID: 29929421; PMCID: PMC6150618.
Sivaramakrishnan P, Gordon AJE, Halliday JA, Herman C. How Acts of Infidelity Promote DNA Break Repair: Collision and Collusion Between DNA Repair and Transcription. Bioessays. 2018 Oct;40(10):e1800045. doi: 10.1002/bies.201800045. Epub 2018 Aug 9. PMID: 30091472; PMCID: PMC6334755.
Sivaramakrishnan P, Sepúlveda LA, Halliday JA, Liu J, Núñez MAB, Golding I, Rosenberg SM, Herman C. The transcription fidelity factor GreA impedes DNA break repair. Nature. 2017 Oct 12;550(7675):214-218. doi: 10.1038/nature23907. Epub 2017 Oct 4. PMID: 28976965; PMCID: PMC5654330.
Satory D, Gordon AJ, Wang M, Halliday JA, Golding I, Herman C. DksA involvement in transcription fidelity buffers stochastic epigenetic change. Nucleic Acids Res. 2015 Dec 2;43(21):10190-9. doi: 10.1093/nar/gkv839. Epub 2015 Aug 24. PMID: 26304546; PMCID: PMC4666387.
Gordon AJ, Satory D, Halliday JA, Herman C. Lost in transcription: transient errors in information transfer. Curr Opin Microbiol. 2015 Apr;24:80-7. doi: 10.1016/j.mib.2015.01.010. Epub 2015 Jan 28. PMID: 25637723; PMCID: PMC4380820.
Gordon AJ, Satory D, Wang M, Halliday JA, Golding I, Herman C. Removal of 8-oxo-GTP by MutT hydrolase is not a major contributor to transcriptional fidelity. Nucleic Acids Res. 2014 Oct 29;42(19):12015-26. doi: 10.1093/nar/gku912. Epub 2014 Oct 7. PMID: 25294823; PMCID: PMC4231768.
Satory D, Halliday JA, Sivaramakrishnan P, Lua RC, Herman C. Characterization of a novel RNA polymerase mutant that alters DksA activity. J Bacteriol. 2013 Sep;195(18):4187-94. doi: 10.1128/JB.00382-13. Epub 2013 Jul 12. PMID: 23852871; PMCID: PMC3754754.
Gordon AJ, Satory D, Halliday JA, Herman C. Heritable change caused by transient transcription errors. PLoS Genet. 2013 Jun;9(6):e1003595. doi: 10.1371/journal.pgen.1003595. Epub 2013 Jun 27. PMID: 23825966; PMCID: PMC3694819.
Satory D, Gordon AJ, Halliday JA, Herman C. Epigenetic switches: can infidelity govern fate in microbes? Curr Opin Microbiol. 2011 Apr;14(2):212-7. doi: 10.1016/j.mib.2010.12.004. PMID: 21496764.
Blankschien MD, Lee JH, Grace ED, Lennon CW, Halliday JA, Ross W, Gourse RL, Herman C. Super DksAs: substitutions in DksA enhancing its effects on transcription initiation. EMBO J. 2009 Jun 17;28(12):1720-31. doi: 10.1038/emboj.2009.126. Epub 2009 May 7. PMID: 19424178; PMCID: PMC2699356.
Gordon AJ, Halliday JA, Blankschien MD, Burns PA, Yatagai F, Herman C. Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 2009 Feb 24;7(2):e44. doi: 10.1371/journal.pbio.1000044. PMID: 19243224; PMCID: PMC2652393.
Blankschien MD, Potrykus K, Grace E, Choudhary A, Vinella D, Cashel M, Herman C. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 2009 Jan;5(1):e1000345. doi: 10.1371/journal.pgen.1000345. Epub 2009 Jan 16. PMID: 19148274; PMCID: PMC2613031.
Horizontal Gene Transfer
Microbes frequently acquire genes from others in a process called horizontal gene transfer (HGT). Many processes can contribute to HGT, including phage-mediated transduction or natural transformation, however, we’ve become interested in a particular process called conjugation. Conjugation is mediated by an autonomous genetic unit that commonly comes in a plasmid or prophage-like form. This genetic unit encodes for a massive cell-envelope mounted secretion system called a Type 4 Secretion System (T4SS). This T4SS is responsible for binding a recipient cell and transferring its own genetic element to the new host.
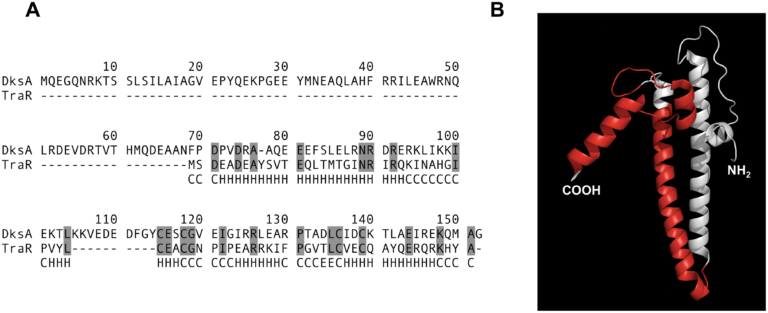
We became interested in these conjugative systems because they have a tendency of interacting with core host processes. The first example of this came when a member of the lab discovered that the most heavily studied conjugative system, F, encoded a dksA homolog called traR. This protein seems to manipulate the transcriptional state of the host cell to favor productive conjugative transfer.
Subsequent to this work, we’ve become interested in studying these systems independent of our other research directions. As mediators of HGT, conjugative systems are key drivers of microbial diversity and evolution, making their presence universally important, may you be a microbiologist or a person with staph infection.
As we move forward, we hope to gain a better understanding of how these systems interface with their hosts and work to spread to broader and broader populations. For more on this subject, please refer to the following publications:
Grace ED, Gopalkrishnan S, Girard ME, Blankschien MD, Ross W, Gourse RL, Herman C. Activation of the σE-dependent stress pathway by conjugative TraR may anticipate conjugational stress. J Bacteriol. 2015 Mar;197(5):924-31. doi: 10.1128/JB.02279-14. Epub 2014 Dec 22. PMID: 25535270; PMCID: PMC4325106.
Blankschien MD, Potrykus K, Grace E, Choudhary A, Vinella D, Cashel M, Herman C. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 2009 Jan;5(1):e1000345. doi: 10.1371/journal.pgen.1000345. Epub 2009 Jan 16. PMID: 19148274; PMCID: PMC2613031.








