Developmental Gliogenesis: Transcriptional Regulation of Glial Cell Fates
A longstanding area of interest in our laboratory is the transcriptional mechanisms governing developmental gliogenesis. As a postdoc, Ben discovered that the transcription factor NFIA regulates the initiation of gliogenesis (Deneen, et al. Neuron 2006) and our lab found that Sox9 and NFIA comprise a transcriptional cascade essential for the gliogenic switch (Kang, et al. Neuron 2012). Additional developmental studies in this area revealed that NFIA and Sox10 mutually antagonize each other’s function and this regulates the generation of the glial sub-lineages, with NFIA being a driver of astrocyte differentiation (Glasgow, et al. Nature Neuroscience 2014).
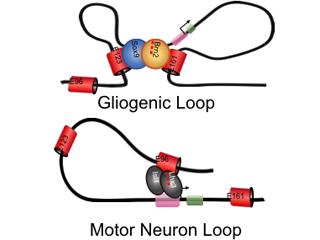
More recently we discovered that lineage specific, three-dimensional chromatin architectures (or chromatin loops) regulate NFIA expression in glial precursors and motor neurons during embryonic development (Glasgow, et al Nature Neuroscience 2017). Currently we are investigating the dynamics of these chromatin loops across development and tumorigenesis, and mapping how changes in epigenetic states at the associated NFIA enhancers regulate chromatin architecture. More broadly, we are delineating how global changes in chromatin looping and epigenetic states influence early glial development.
Astrocyte Diversity: New Perspectives and Approaches
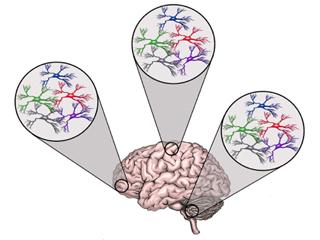
Astrocytes play diverse roles in all facets of brain physiology, suggesting the existence of an extensive reservoir of diverse subpopulations in the adult brain. In fact, Cajal initially described extensive morphological heterogeneity of astrocyte populations in the brain over 100 years ago. Since then, our understanding of the molecular and cellular heterogeneity of astrocytes has remained stagnant, with astrocytes traditionally grouped into two broad categories: fibrous and protoplasmic.
While we were the first to show that patterning oversees the generation of diverse astrocytes in the embryonic spinal cord (Hochstim, et al. Cell 2008), applying these principles to astrocyte populations in the adult brain has remained a challenge because it requires an intimate knowledge of region-specific patterning mechanisms and the associated mouse tools to gain access to these populations. To overcome these limitations we have developed novel tactics and mouse tools that serve as an entry point for unlocking the nature of astrocyte diversity in the brain (Lin, et al. Nature Neuroscience 2017). Using these tools and tactics, we identified several astrocyte subpopulations that are endowed with distinct cellular, molecular, and functional properties in the adult brain.
Gliogenesis and Malignant Glioma: Crossroads of Cell Fate and Malignancy
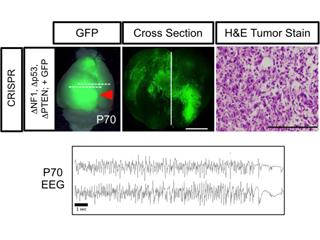
Two critical intersection points for the mechanisms underlying glial development are glioma tumorigenesis and white matter injury. To better understand the enigmatic nature of glioma biology, it is critical to decipher how the mechanisms that drive glial development are utilized during tumorigenesis. We have focused our studies on the role of NFIA in glioma tumorigenesis, finding that it plays a critical role in glioma tumorigenesis (Glasgow, et al. Journal of Neuroscience 2013) and can influence the generation of glioma subtypes (Glasgow, et al. Nature Neuroscience 2014).
In a separate series of studies, we identified diverse glioma subpopulations whose emergence during tumor progression is linked to glioma-induced epilepsy (Lin, et al. Nature Neuroscience 2017). This unexpected finding has paved the way for a series of studies aimed at delineating how glioma subpopulations influence changes in brain circuitry and how neuronal activity influences glioma progression.
CNS Injury and Repair: Glial Insights into Repairing the Brain
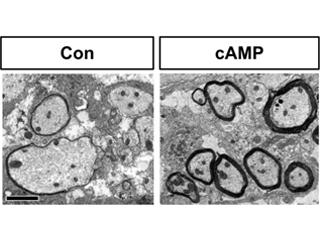
White matter injury (WMI) produces a cascade of cellular responses that contribute to both pathogenesis and recovery, including the generation of reactive astrocytes. Using our developmental studies on astrocyte lineage as an entry point, we identified a novel cohort of genes expressed in human reactive astrocytes and further defined the role of Asef in blood-brain-barrier (BBB) remodeling after WMI (Chaboub, et al. Journal of Neuroscience 2016).
In a separate series of studies, we have begun to study how gliogenic developmental factors influence WMI responses. We discovered that the Wnt-modifying gene, Daam2, suppresses repair after adult and neonatal WMI (Lee, et al. Neuron 2015). More recently, we have studied the role of NFIA and its associated biology in reactive astrocytes after both WMI and cortical stroke, finding that it has distinct functional mechanisms of action in different injury models.
Functional Genomics: Leveraging Mouse Models of Glioma
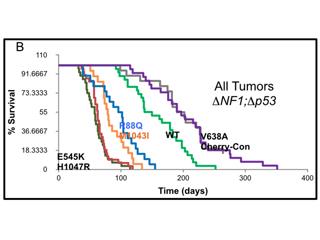
In the era of personalized medicine, significant efforts are being made to comprehensively characterize the spectrum of mutations associated with a given disease. In order for these efforts to be predictive of outcomes, it is essential to decipher the functional significance of disease associated mutations. Towards this goal, in collaboration with Ken Scott’s group we recently developed DNA barcoding based methodologies that allow us to screen the function of up to 50 gene variants at a time in our glioma model. Capitalizing on the genetic flexibility of our mouse models of glioma, we are performing in vivo complementation screening of all known variants of key oncogenic drivers in malignant glioma. In parallel studies, in collaboration with cancer epidemiologist Melissa Bondy, we are we are functionally examining a cohort of susceptibility variants linked to familial glioma.
Contact
Benjamin Deneen, Ph.D.
Principal Investigator
deneen@bcm.edu
Saritha Krishna, Ph.D.
Lab Manager
saithakn@bcm.edu








