Macromolecular structure and dynamics can be probed with a wide variety of multi-dimensional and multi-nuclear NMR methods that can be implemented on our 600 and 800 MHz Bruker NMR spectrometers. Protein sequential assignment and structure determination usually require stable isotope labeling with 13C and 15N, and sometimes 2H, but inexpensive 15N labeling can be used for pilot experiments to determine the feasibility of structural biology projects such as structure determination or macromolecule-observed mapping of ligand binding. 2D 1H/15N HSQC (or TROSY) spectra acquired in a few minutes or hours, depending on concentration, show peaks for every backbone and sidechain amide proton. Well dispersed backbone amide peaks (one for each non-proline residue), such as the black peaks in Figure 1, show that the sample is well-behaved and amenable to study.
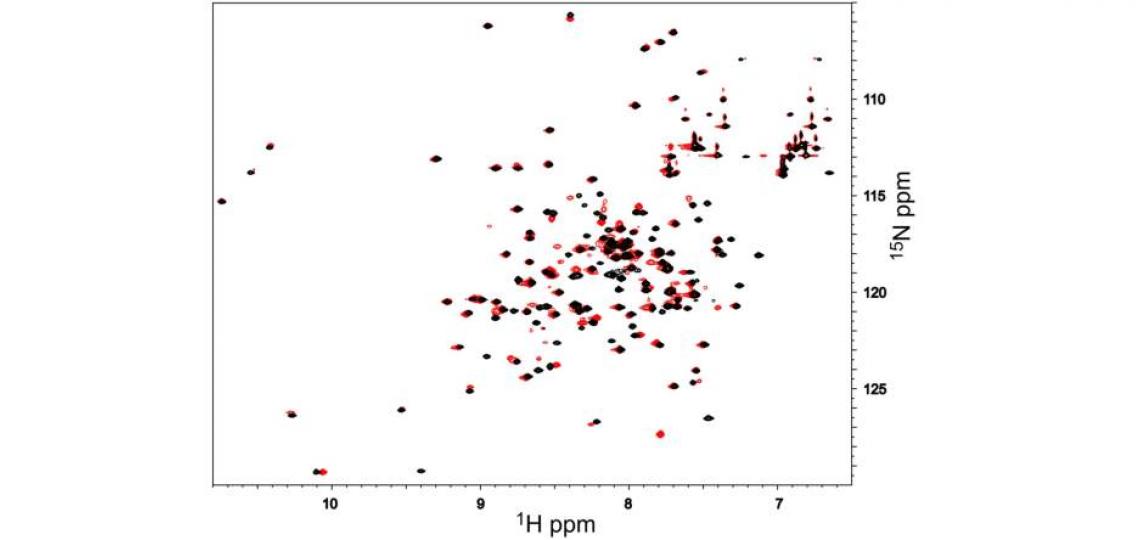
Figure 1: 2D 1H-15N HSQC spectra of 1 mM androcam protein alone (black) and with an excess of an unlabeled peptide derived from myosin VI (red). Each 1H/15N peak corresponds to one backbone or side chain amide. Perturbations of only a subset of peaks (e.g. from 7.45/126.5 in black to 7.78/127.4 in red) show that peptide binding affects only some parts of the labeled protein. To map these, the residues corresponding to the peaks in the spectrum must be identified. For many proteins or domains of interest, chemical shift assignments are available from the literature. De novo determination of the backbone atom chemical shifts for this 18 kDa protein required 15N/13C labeling and several days of NMR time. This work was the first step in an NMR structure determination effort.
This protein, androcam, is known to bind a target peptide; addition of excess peptide and re-acquisition of the spectrum shows (Figure 1, red peaks) that a subset of backbone amides experience chemical shift perturbations (CSPs) upon peptide binding. Perturbed peaks are near the peptide binding site. Sequential assignment of the resonances allows the identity of each peak in the 2D spectrum to be known, so that the binding site can be mapped onto the structure. These chemical shift perturbations map to the C-terminal domain of androcam (a calmodulin homolog); the resonances of the N-terminal lobe are unperturbed. However, weak binding of Ca2+ or Mg2+ does alter the chemical shifts of amides in the N-terminal lobe as revealed by NMR titrations (Figure 2).
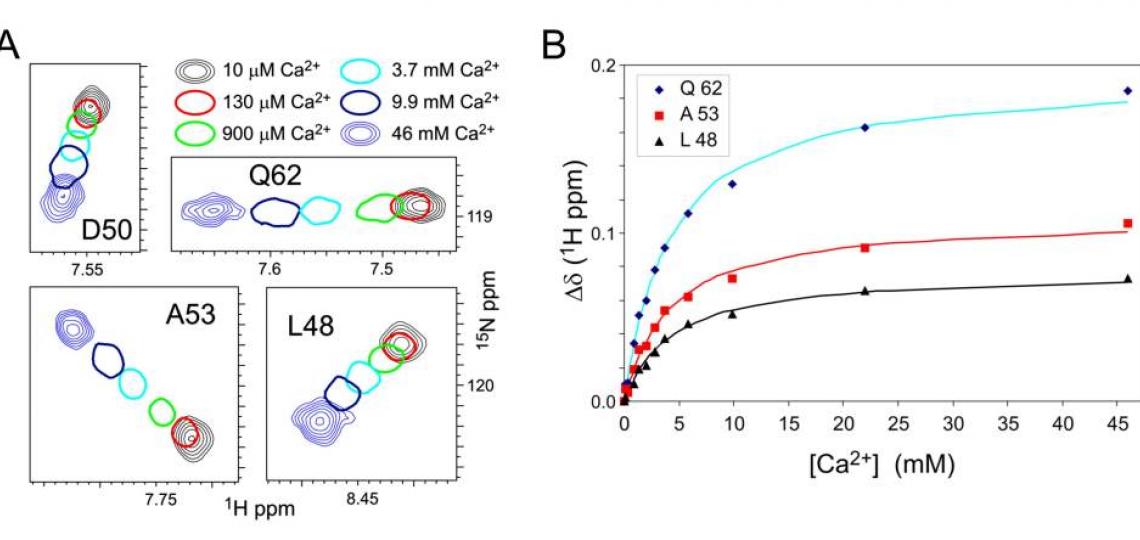
Figure 2. A. Regions of 1H-15N HSQC spectra of 1 mM androcam protein at different free CaCl2concentrations. For intermediate [CaCl2], only one contour is drawn. Most androcam backbone amide peaks are unaffected by [CaCl2] in this range, but some change in a saturable manner that reflect a weak binding event. These CSPs map exclusively to the N-terminal lobe, either in or adjacent to a novel metal binding site that comprises residues from two defunct EF-hand domains. B. Chemical shift perturbations (CSPs) for androcam on CaCl2 titration are least-squares fit to a binding constant KD ~ 4 mM. Titration with MgCl2 gives similar CSPs and KD, indicating that the site binds divalent metals with little specificity. (The closely related protein calmodulin, 65% identity, shows a strong preference for Ca2+over Mg2+ at all four of its EF-hand metal binding sites.) The lack of metal specificity shows that the androcam N-terminal lobe has lost the “calcium switch” properties of the homolog calmodulin from which it evolved.
Macromolecular CSPs can report on conformational averaging between free and bound states, and so can be used to extract binding constants for (weakly binding) ligands, as shown in Figure 2B. The analysis does not require the peak identities to be known. CSP analysis as a follow-up to ‘ligand-observed’ binding methods such as saturation transfer difference NMR or Water-LOGSY could help determine if different ligands bind to the same or different sites, even without backbone resonance assignments.
Although the testis-specific protein androcam binds the myosin VI target peptide with only its C-terminal domain, the ubiquitous calcium adapter protein calmodulin binds the same peptide with both lobes (Figure 3). Well folded proteins like androcam experience modest, localized chemical shift perturbations upon binding to partners that can be used to identify the binding surface; the dramatic chemical shift perturbations seen for almost every calmodulin residue reflect co-folding or global conformational changes.
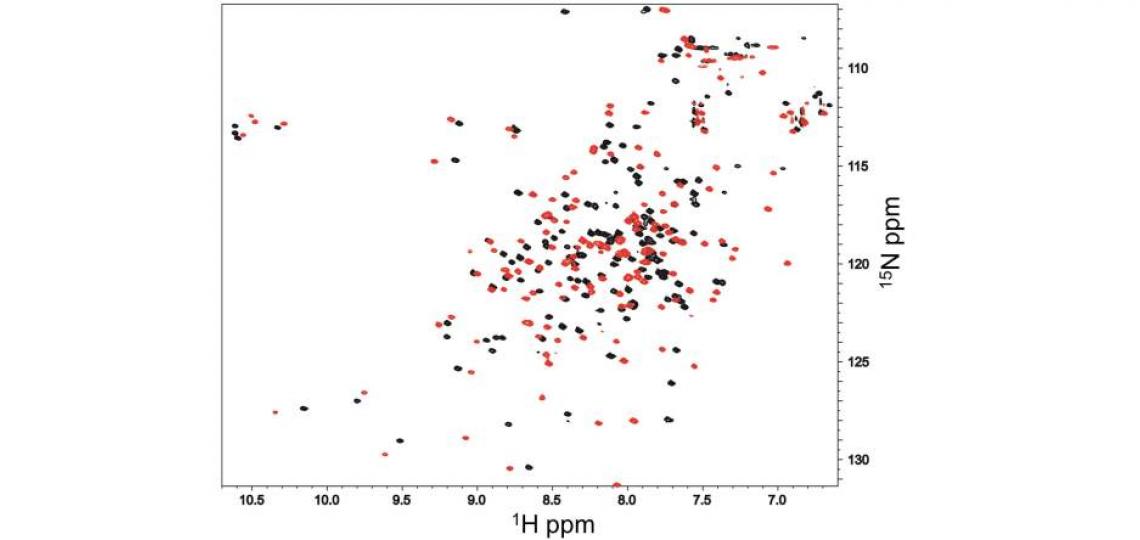
Figure 3: 1H-15N HSQC spectra of 1 mM calmodulin/CaCl2 alone (black) and in the presence of an excess of an unlabeled peptide derived from myosin VI (red). Almost every calmodulin 1H/15N peak exhibits substantial chemical shift perturbation, indicating that the whole calmodulin structure is dramatically reorganized by peptide binding, in contrast with what is seen for androcam (Figure 1).
Macromolecular NMR methods such as these can be applied in a directed way to test detailed hypotheses (e.g. does the peptide binds to one lobe or both?) that build on biochemical or structural evidence from other methods. The types of 2D spectra shown here, even without sequential assignment of the resonances, would be excellent preliminary data for a grant to justify further experiments to explore the mapping of a ligand binding site or the nature of a conformational reorganization induced by ligand binding. Part of the proposed future work might include assignment of the backbone resonances (de novo or from reported literature) in order to map the ligand-induced chemical shift changes to regions of the primary and 3D structure.
References:
C Aguirre, O Cala, I Krimm, Overview of Probing Protein-Ligand Interactions Using NMR, Curr Protoc Protein Sci, 2015, 81 (17.18), pp. 1-24.
MK Joshi, S Moran, KM Beckingham, and KR MacKenzie, Structure of androcam supports specialized interactions with myosin VI, Proc Natl Acad Sci USA, 2012, 109, pp 13290-5.
Contacts:
Kevin MacKenzie, Ph.D.
Kevin.MacKenzie@bcm.edu
281-773-2672








