About the Lab
RNAs have rapidly emerged as the key players in many fundamental biological processes, as well as the bridge to future medicine. The Yen laboratory is interested in the biology of RNA molecular switches (riboswitch), small catalytic RNAs, mutational RNA events in disease, and their applications in medicine. We use a variety of chemical and molecular biology strategies, in combination with mammalian cell cultures and animal models in pursuit of our goals. View information about our research areas below.
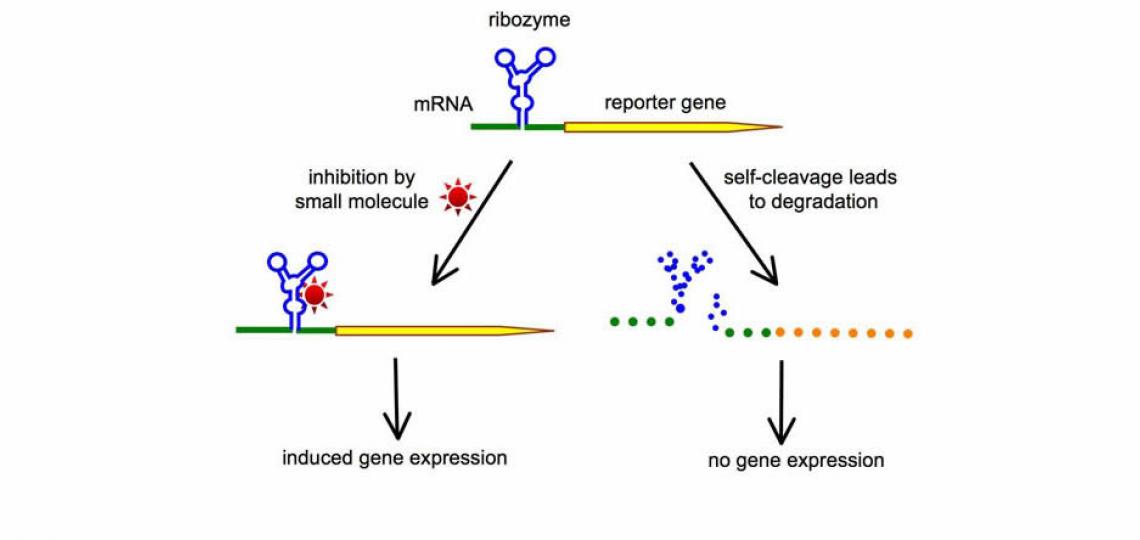
Development of RNA-based Switches for Gene Regulation
The ability to control gene expression has always been indispensable in order to elucidate the function of a specific gene product, or to manipulate the levels of a specific protein to achieve therapeutic effects. A major focus of our lab is to harness the power of small catalytic RNAs (such as the hammerhead ribozyme) to create new RNA switches that can be turned on or off by small molecules. One of the strategies we use is illustrated in Figure 1. When a self-cleaving ribozyme is embedded in the mRNA, the spontaneous self-cleavage of ribozyme leads to destruction of the mRNA and therefore a loss of gene expression. Small molecules capable of inhibiting ribozyme result in preservation of the intact mRNA, and therefore induce gene expression. This strategy of RNA switches has the potential to lead to the creation of many tailor-made gene regulation systems, each controlled by a different small molecule. An important goal of our future research is to develop conditional RNA switches that can be turned on or off by FDA-approved small molecules. Such gene regulation systems, combining safe small molecules with non-immunogenic RNA switches, would be significantly safer to use in clinical applications as well as in biological studies.
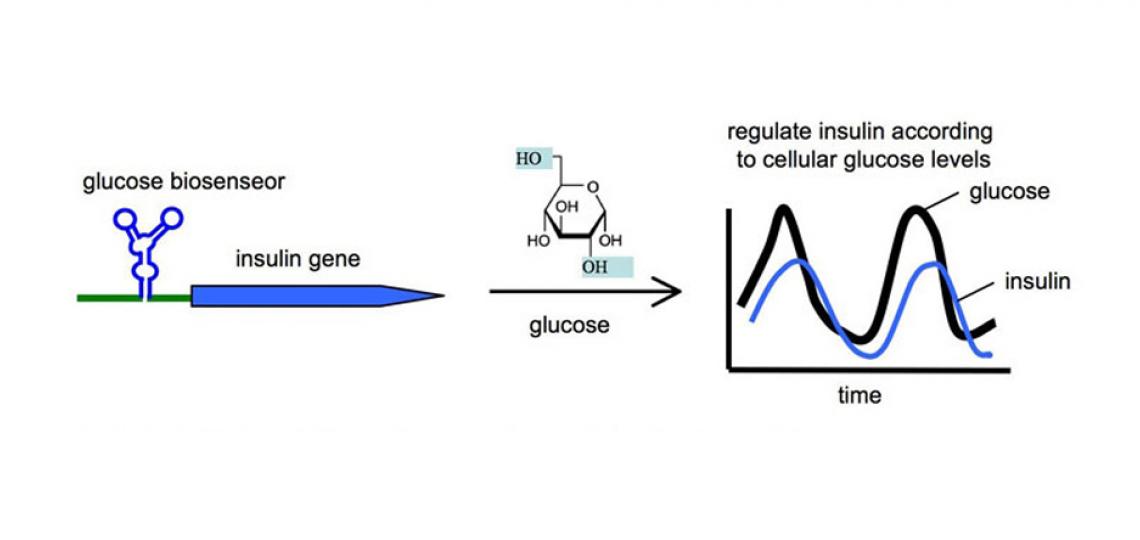
Engineering RNA-based Molecular Biosensors for Ligand Detection
The small molecules used to control the RNA switch can be any cellular biomolecule, including metabolites, siRNA, miRNA, or proteins. In fact, it has been possible to engineer the ribozyme as a sensor to recognize a specific cellular ligand, and turn on a transgene according to the levels of the cellular ligand. We have great interest in exploring the engineering principles required to create sophisticated RNA switches that function as biosensors in vivo. Such biosensors would provide spatial as well as temporal information regarding the levels of specific ligands in disease, and the input information can be used to regulate cellular behavior for achieving therapeutic goals. For example, an RNA biosensor can be engineered to recognize glucose as its ligand, and in response, regulates the expression of an engineered insulin protein to modulate the glucose levels in diabetic patients (Figure 2). Similar biosensor platforms can also function as safety switches. For example, a biosensor can be engineered to detect the presence of a cancer biomarker in stem cells. When a normal stem cell erroneously transforms into a cancer cell, the biosensor would switch on a suicidal gene for self-destruction.
Investigating RNA Mutations in Human Cancer for Diagnosis and Therapy
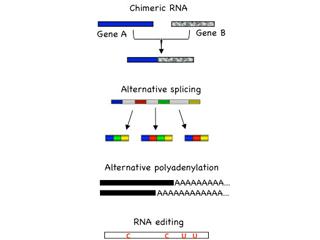
Transcription-induced RNA mutations include chimeric RNA, alternative splicing, alternative polyadenylation, and RNA editing. The prevalence of RNA mutations may allow the limited number of human genes to encode substantially larger number of RNAs and proteins, forming an additional layer of cellular complexity. In-depth analysis of RNA mutations in cancer has the potential to illuminate the underlying mechanisms of cancer formation, as well as develop useful tools for diagnosis and therapy. One of our focuses is to identify key driver RNA mutations in human cancer using a combined strategy of high-throughput sequencing, bioinformatics, mammalian cell cultures and animal models. Such RNA mutations, once identified, could serve as biomarkers for early cancer detection, as well as provide drug targets for cancer therapy.








