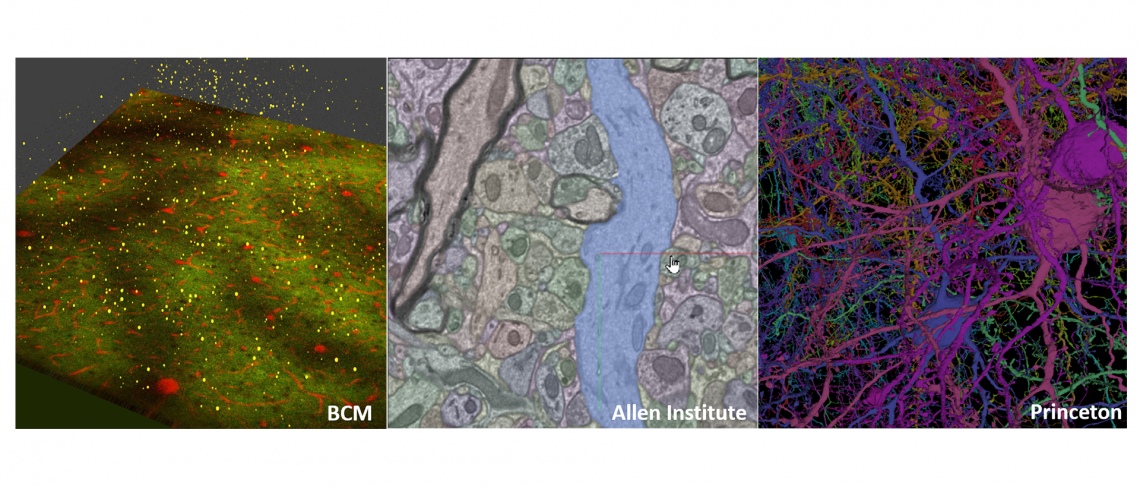
Detailed, 3D map created from mouse brain to support machine learning
A 3D wiring diagram, the largest of its kind, containing hundreds of thousands of cells and nearly half a billion connections of a mouse brain has been created by researchers at Baylor College of Medicine, the Allen Institute and Princeton University as part of a project to learn principles from the brain that can help advance artificial intelligence. This unique resource was generated through the IARPA Machine Intelligence from Cortical Networks (MICrONS) program, and will be invaluable for neuroscientists seeking to understand how the brain processes information along neocortical circuits, and for researchers wanting to treat brain disorders where wiring or connections are altered.
The dataset took five years to create and is publicly available for anyone in the community to browse and use. It maps the fine structures and connectivity of approximately 200,000 brain cells and nearly 500 million synapses, all contained in a cubic millimeter chunk of mouse brain approximately the size of a grain of sand. The area of the brain that was chosen is from the visual neocortex, a part of the brain that is essential for visual perception. A unique feature of the dataset is that researchers at Baylor College of Medicine collected comprehensive recordings of the activity patterns evoked by a variety of complex visual stimuli, from YouTube clips to Hollywood movies, for around 75,000 brain cells in the same brain volume that was used to generate the nano-scale connectivity map.
“It’s been a privilege to work on what has truly been a moon-shot project,” said Paul Fahey, postdoctoral associate at Baylor. “Every step of this collaboration has pushed the limits of our technical capabilities, and the result is something very special.”
The anatomical data was collected using serial section electron microscopy (EM), which reveals incredible details of an entire microscopic landscape, including the densely packed topography of tendrilled neurons, astrocytes, blood vessels and other cells, as well as all the miniscule internal components of each cell. The dataset joins the ranks of other recently released wiring diagrams from human and fruit fly brains described using this technique.
"The neocortex contains billions of neurons communicating through trillions of connections that have endowed mammals with astonishing capabilities like perception, cognition and motor control. A key question to untangling this bewildering complexity is to discover the relationships between the wiring rules and the functional properties of neurons,” said Dr. Andreas Tolias, professor of neuroscience and director of the Center for Neuroscience and Artificial Intelligence at Baylor and one of the lead scientists of MICrONS. “This program is unique since it enabled us to assemble an interdisciplinary team to perform a very ambitious experiment that will allow us to address this question.”
Before the EM imaging and cellular reconstruction was performed by researchers at the Allen Institute and Princeton, the research team at Baylor measured the activity of neurons as the mouse viewed images or movies of natural scenes.
“This was a relay race that spanned three institutions across the country over several years,” said Dr. Jacob Reimer, assistant professor of neuroscience at Baylor. “We had the baton first. While we were collecting the functional imaging data, we were constantly aware of the extraordinary effort that our colleagues at Allen and Princeton were about to put into understanding the anatomy of this same volume of tissue.”
After the Baylor experiments, Allen Institute researchers preserved and sliced the brain piece in more than 27,000 thin slices – each no more than 1/400th the thickness of a human hair – and then captured a total of 150 million images of those slices using customized electron microscopes. The research team at Princeton then used novel machine learning approaches to segment the images, defining each cell and its internal components individually.
The end result: intricate digital renderings of 200,000 brain cells and the connections between them, many of which have been matched back to the dynamic activity patterns that were recorded as they participated in visual processing in the living brain.
“We’re basically treating a brain circuit as a computer, and we asked three questions: What does it do? How is it wired up? What is the program?” said Dr. R. Clay Reid, senior investigator at the Allen Institute and one of the lead scientists of MICrONS. “Experiments were done to literally see the neurons’ activity, to watch them compute. The reconstructions that we’re presenting today let us see the elements of the neural circuit: the brain cells and the wiring, with the ability to follow the wires to map the connections between cells. The final step is to interpret this network, at which point we may be able to say we can read the brain’s program.”
“Computer programs have gone through a massive revolution in the past decade, but something is still missing: today’s programs are easy to fool and they generalize poorly to new situations. In many ways they are much dumber than a mouse,” said Dr. Xaq Pitkow, associate professor of neuroscience and a McNair Scholar at Baylor, assistant professor of electrical and computer engineering at Rice University and one of the lead scientists of MICrONS. “Our new exquisitely detailed data about the mouse brain gives us a chance to find what makes the biological brain smarter. And that could help us develop smarter computers.”
The wiring diagram contains the most cells and connections of any such dataset to date, and it is large enough to capture entire local circuits and near-complete 3D shapes of individual mouse neurons.
Our five-year mission had an ambitious goal that many regarded as unattainable. Today, we have been rewarded by breathtaking new vistas of the mammalian cortex," said Dr. H. Sebastian Seung, professor of neuroscience and computer science at Princeton University and one of the lead scientists of MICrONS. "As we transition to a new phase of discovery, we are building a community of researchs to use the data in new ways. Only the collective efforts of this community can realize the resources's potential for enabling new discoveries about the brain."
The project is supported by the Intelligence Advanced Research Projects Activity (IARPA) via Department of Interior / Interior Business Center (DoI/IBC) under contracts D16PC00003 (Baylor), D16PC00004 (Allen), and D16PC0005 (Princeton). The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright annotation thereon. Disclaimer: The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of IARPA, DoI/IBC, or the U.S. Government.
MICrONS is hosted online by the Brain Observatory Storage Service & Database with cloud-data-storage support from Google and Amazon.










 Credit
Credit