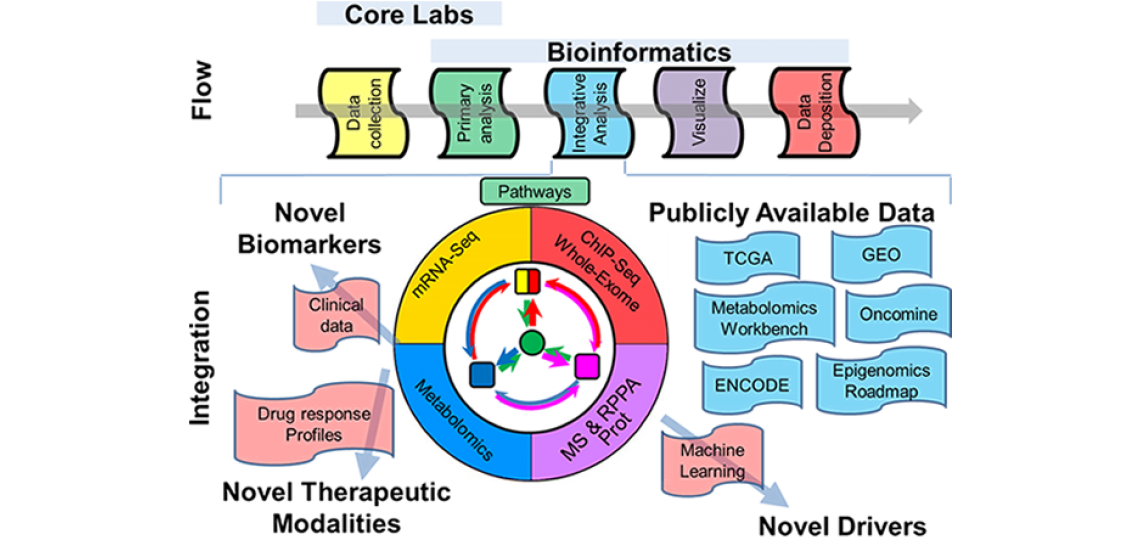
Overview
The Multi-Omics Data Analysis group assists with analysis of data generated by the major technology platforms of the CPRIT Core Facility, including mass spectrometry (MS) metabolomics, MS-based proteomics and reverse phase protein array (RPPA) proteomics. In addition, we accept and processes high-throughput sequencing data including:
- Coding and noncoding transcriptomics: RNA-Seq, smallRNA-Seq
- Single cell transcriptomics scRNA-Seq
- Cistrome: ChIP-Seq, Reduced Representation Bisulfite Sequencing (RRBS), and Whole Genome Bisulfite Sequencing (WGBS)
- Genomics: Whole Genome Sequencing (WGS) or Whole Exome sequencing (WES)
Using a consistent data work-flow, we assist with Primary/Tier 1 data analysis in close collaboration with the Core Facility leadership. Additionally, we provide as independent support services Integrative/Tier 2 analysis for multiple omics data to lead to systems biology level insight, and also to generate robust testable hypotheses. Integrative analysis can be further performed using data sets from national and international projects, such as The Cancer Genome Atlas (TCGA), Encode, the NIH Epigenomic Roadmap, International Human Epigenome Consortium (IHEC), The Metabolomics Workbench, as well as data sets from scientific community repositories such as the NIH Gene Expression Omnibus (GEO) and NIH Short Read Archive (SRA).
References (Core Supported Publications)
Multi-Omics Data Analysis
Dan L Duncan Comprehensive Cancer Center
Baylor College of Medicine
One Baylor Plaza
Suite 450A
Houston, TX 77030








